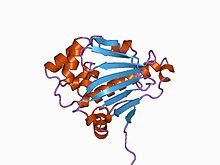
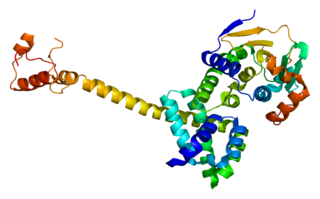In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to assist large proteins in proper protein folding during or after synthesis, and after partial denaturation. Chaperones are also involved in the translocation of proteins for proteolysis.
Hsp90 is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs.
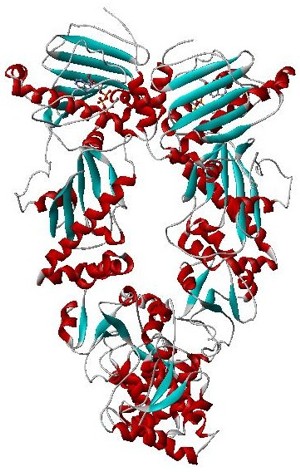
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms.

Heat shock protein HSP 90-alpha is a protein that in humans is encoded by the HSP90AA1 gene.
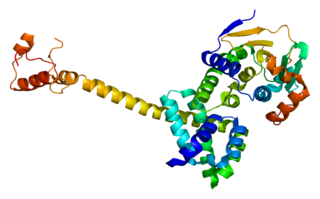
Hsp90 co-chaperone Cdc37 is a protein that in humans is encoded by the CDC37 gene. This protein is highly similar to Cdc 37, a cell division cycle control protein of Saccharomyces cerevisiae. This protein is a HSP90 Co-chaperone with specific function in cell signal transduction. It has been shown to form complex with Hsp90 and a variety of protein kinases including CDK4, CDK6, SRC, RAF1, MOK, as well as eIF-2 alpha kinases. It is thought to play a critical role in directing Hsp90 to its target kinases.

Heat shock protein HSP 90-beta also called HSP90beta is a protein that in humans is encoded by the HSP90AB1 gene.

Prostaglandin E synthase 3 (cytosolic) is an enzyme that in humans is encoded by the PTGES3 gene.

AH receptor-interacting protein (AIP) also known as aryl hydrocarbon receptor-interacting protein, immunophilin homolog ARA9, or HBV X-associated protein 2 (XAP-2) is a protein that in humans is encoded by the AIP gene. The protein is a member of the FKBP family.

Hsc70-interacting protein also known as suppression of tumorigenicity 13 (ST13) is a protein that in humans is encoded by the ST13 gene.

BAG family molecular chaperone regulator 2 is a protein that in humans is encoded by the BAG2 gene.

Heat shock protein 75 kDa, mitochondrial is a protein that in humans is encoded by the TRAP1 gene.

Mitochondrial import receptor subunit TOM34 is a protein that in humans is encoded by the TOMM34 gene.

BAG family molecular chaperone regulator 4 is a protein that in humans is encoded by the BAG4 gene.

ATP synthase subunit e, mitochondrial is an enzyme that in humans is encoded by the ATP5ME gene.
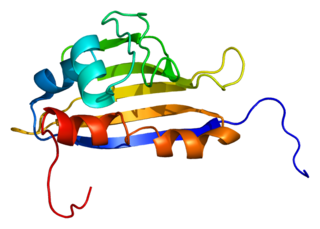
Activator of 90 kDa heat shock protein ATPase homolog 1 is an enzyme that in humans is encoded by the AHSA1 gene.
AHSA2 also known as AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) is a human gene which encodes a protein which acts as co-chaperone of Hsp90. AHSA2 and the related AHSA1 belongs to the AHA family of stress-regulated proteins that bind directly to Hsp90 and are required for Hsp90-dependent activation of client proteins.

An Hsp90 inhibitor is a substance that inhibits that activity of the Hsp90 heat shock protein. Since Hsp90 stabilizes a variety of proteins required for survival of cancer cells, these substances may have therapeutic benefit in the treatment of various types of malignancies. Furthermore, a number of Hsp90 inhibitors are currently undergoing clinical trials for a variety of cancers. Hsp90 inhibitors include the natural products geldanamycin, Retaspimycin hydrochloride and radicicol as well as semisynthetic derivatives 17-N-Allylamino-17-demethoxygeldanamycin (17AAG).
The chaperone code refers to post-translational modifications of molecular chaperones that control protein folding. Whilst the genetic code specifies how DNA makes proteins, and the histone code regulates histone-DNA interactions, the chaperone code controls how proteins are folded to produce a functional proteome.

Folliculin-interacting protein 1 (FNIP1) functions as a co-chaperone which inhibits the ATPase activity of the chaperone Hsp90 and decelerates its chaperone cycle. FNIP1 acts as a scaffold to load FLCN onto Hsp90. FNIP1 is also involved in chaperoning of both kinase and non-kinase clients.