In computer science, string-searching algorithms, sometimes called string-matching algorithms, are an important class of string algorithms that try to find a place where one or several strings are found within a larger string or text.
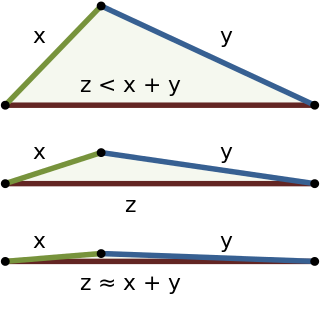
In mathematics, the triangle inequality states that for any triangle, the sum of the lengths of any two sides must be greater than or equal to the length of the remaining side. This statement permits the inclusion of degenerate triangles, but some authors, especially those writing about elementary geometry, will exclude this possibility, thus leaving out the possibility of equality. If x, y, and z are the lengths of the sides of the triangle, with no side being greater than z, then the triangle inequality states that

The longest common subsequence (LCS) problem is the problem of finding the longest subsequence common to all sequences in a set of sequences. It differs from the longest common substring problem: unlike substrings, subsequences are not required to occupy consecutive positions within the original sequences. The longest common subsequence problem is a classic computer science problem, the basis of data comparison programs such as the diff utility, and has applications in computational linguistics and bioinformatics. It is also widely used by revision control systems such as Git for reconciling multiple changes made to a revision-controlled collection of files.
In computational linguistics and computer science, edit distance is a string metric, i.e. a way of quantifying how dissimilar two strings are to one another, that is measured by counting the minimum number of operations required to transform one string into the other. Edit distances find applications in natural language processing, where automatic spelling correction can determine candidate corrections for a misspelled word by selecting words from a dictionary that have a low distance to the word in question. In bioinformatics, it can be used to quantify the similarity of DNA sequences, which can be viewed as strings of the letters A, C, G and T.
In computer science, the Rabin–Karp algorithm or Karp–Rabin algorithm is a string-searching algorithm created by Richard M. Karp and Michael O. Rabin (1987) that uses hashing to find an exact match of a pattern string in a text. It uses a rolling hash to quickly filter out positions of the text that cannot match the pattern, and then checks for a match at the remaining positions. Generalizations of the same idea can be used to find more than one match of a single pattern, or to find matches for more than one pattern.
In computer science, the Boyer–Moore string-search algorithm is an efficient string-searching algorithm that is the standard benchmark for practical string-search literature. It was developed by Robert S. Boyer and J Strother Moore in 1977. The original paper contained static tables for computing the pattern shifts without an explanation of how to produce them. The algorithm for producing the tables was published in a follow-on paper; this paper contained errors which were later corrected by Wojciech Rytter in 1980. The algorithm preprocesses the string being searched for, but not the string being searched in. It is thus well-suited for applications in which the pattern is much shorter than the text or where it persists across multiple searches. The Boyer–Moore algorithm uses information gathered during the preprocess step to skip sections of the text, resulting in a lower constant factor than many other string search algorithms. In general, the algorithm runs faster as the pattern length increases. The key features of the algorithm are to match on the tail of the pattern rather than the head, and to skip along the text in jumps of multiple characters rather than searching every single character in the text.

In computer science, a suffix tree is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
In computer science, a suffix array is a sorted array of all suffixes of a string. It is a data structure used in, among others, full-text indices, data-compression algorithms, and the field of bibliometrics.
The Lenstra–Lenstra–Lovász (LLL) lattice basis reduction algorithm is a polynomial time lattice reduction algorithm invented by Arjen Lenstra, Hendrik Lenstra and László Lovász in 1982. Given a basis with n-dimensional integer coordinates, for a lattice L with , the LLL algorithm calculates an LLL-reduced lattice basis in time
In computer science, the Boyer–Moore–Horspool algorithm or Horspool's algorithm is an algorithm for finding substrings in strings. It was published by Nigel Horspool in 1980 as SBM.
In computer science, the longest common substring problem is to find a longest string that is a substring of two or more strings. The problem may have multiple solutions. Applications include data deduplication and plagiarism detection.
In information theory and computer science, the Damerau–Levenshtein distance is a string metric for measuring the edit distance between two sequences. Informally, the Damerau–Levenshtein distance between two words is the minimum number of operations required to change one word into the other.

In computer science, approximate string matching is the technique of finding strings that match a pattern approximately. The problem of approximate string matching is typically divided into two sub-problems: finding approximate substring matches inside a given string and finding dictionary strings that match the pattern approximately.
BLEU is an algorithm for evaluating the quality of text which has been machine-translated from one natural language to another. Quality is considered to be the correspondence between a machine's output and that of a human: "the closer a machine translation is to a professional human translation, the better it is" – this is the central idea behind BLEU. BLEU was one of the first metrics to claim a high correlation with human judgements of quality, and remains one of the most popular automated and inexpensive metrics.
In computer science and statistics, the Jaro–Winkler distance is a string metric measuring an edit distance between two sequences. It is a variant proposed in 1990 by William E. Winkler of the Jaro distance metric.
In data analysis, cosine similarity is a measure of similarity between two sequences of numbers. For defining it, the sequences are viewed as vectors in an inner product space, and the cosine similarity is defined as the cosine of the angle between them, that is, the dot product of the vectors divided by the product of their lengths. It follows that the cosine similarity does not depend on the magnitudes of the vectors, but only on their angle. The cosine similarity always belongs to the interval For example, two proportional vectors have a cosine similarity of 1, two orthogonal vectors have a similarity of 0, and two opposite vectors have a similarity of -1. The cosine similarity is particularly used in positive space, where the outcome is neatly bounded in .
In mathematics, a covering number is the number of spherical balls of a given size needed to completely cover a given space, with possible overlaps. Two related concepts are the packing number, the number of disjoint balls that fit in a space, and the metric entropy, the number of points that fit in a space when constrained to lie at some fixed minimum distance apart.
In machine learning and data mining, a string kernel is a kernel function that operates on strings, i.e. finite sequences of symbols that need not be of the same length. String kernels can be intuitively understood as functions measuring the similarity of pairs of strings: the more similar two strings a and b are, the higher the value of a string kernel K(a, b) will be.
Information distance is the distance between two finite objects expressed as the number of bits in the shortest program which transforms one object into the other one or vice versa on a universal computer. This is an extension of Kolmogorov complexity. The Kolmogorov complexity of a single finite object is the information in that object; the information distance between a pair of finite objects is the minimum information required to go from one object to the other or vice versa. Information distance was first defined and investigated in based on thermodynamic principles, see also. Subsequently, it achieved final form in. It is applied in the normalized compression distance and the normalized Google distance.

In computer science, a suffix automaton is an efficient data structure for representing the substring index of a given string which allows the storage, processing, and retrieval of compressed information about all its substrings. The suffix automaton of a string is the smallest directed acyclic graph with a dedicated initial vertex and a set of "final" vertices, such that paths from the initial vertex to final vertices represent the suffixes of the string.























