A halophile is an extremophile that thrives in high salt concentrations. In chemical terms, halophile refers to a Lewis acidic species that has some ability to extract halides from other chemical species.

The Codex Alexandrinus, designated by the siglum A or 02, δ 4, is a manuscript of the Greek Bible, written on parchment. Using the study of comparative writing styles (palaeography), it has been dated to the fifth century. It contains the majority of the Greek Old Testament and the Greek New Testament. It is one of the four Great uncial codices. Along with Codex Sinaiticus and Vaticanus, it is one of the earliest and most complete manuscripts of the Bible.
The oxidase test is used to determine if an organism possesses the cytochrome c oxidase enzyme. The test is used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species. It is also used to differentiate pseudomonads from related species.
The Book of Odes is a book of the Bible found only in Eastern Orthodox Bibles and included or appended after the Psalms in Alfred Rahlfs' critical edition of the Septuagint, coming from the fifth-century Codex Alexandrinus. The chapters are prayers and songs (canticles) from the Old and New Testaments. The first nine of them form the basis for the canon sung during matins and other services.
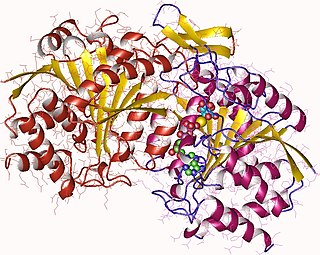
Isocitrate dehydrogenase (NAD+) (EC 1.1.1.41, isocitric dehydrogenase, beta-ketoglutaric-isocitric carboxylase, isocitric acid dehydrogenase, NAD dependent isocitrate dehydrogenase, NAD isocitrate dehydrogenase, NAD-linked isocitrate dehydrogenase, NAD-specific isocitrate dehydrogenase, NAD isocitric dehydrogenase, isocitrate dehydrogenase (NAD), IDH (ambiguous), nicotinamide adenine dinucleotide isocitrate dehydrogenase) is an enzyme with systematic name isocitrate:NAD+ oxidoreductase (decarboxylating). This enzyme catalyses the following chemical reaction

Haloarchaea are a class of the Euryarchaeota, found in water saturated or nearly saturated with salt. Halobacteria are now recognized as archaea rather than bacteria and are one of the largest groups. The name 'halobacteria' was assigned to this group of organisms before the existence of the domain Archaea was realized, and while valid according to taxonomic rules, should be updated. Halophilic archaea are generally referred to as haloarchaea to distinguish them from halophilic bacteria.

In taxonomy, Haloferax is a genus of the Haloferacaceae.
Halocins are bacteriocins produced by halophilic Archaea and a type of archaeocin.
An archaeosortase is a protein that occurs in the cell membranes of some archaea. Archaeosortases recognize and remove carboxyl-terminal protein sorting signals about 25 amino acids long from secreted proteins. A genome that encodes one archaeosortase may encode over fifty target proteins. The best characterized archaeosortase target is the Haloferax volcanii S-layer glycoprotein, an extensively modified protein with O-linked glycosylations, N-linked glycosylations, and a large prenyl-derived lipid modification toward the C-terminus. Knockout of the archaeosortase A (artA) gene, or permutation of the motif Pro-Gly-Phe (PGF) to Pro-Phe-Gly in the S-layer glycoprotein, blocks attachment of the lipid moiety as well as blocking removal of the PGF-CTERM protein-sorting domain. Thus archaeosortase appears to be a transpeptidase, like sortase, rather than a simple protease.

Haloferax volcanii is a species of organism in the genus Haloferax in the Archaea.
Haloferax larsenii is a gram-negative, aerobic, neutrophilic, extremely halophilic archaeon. It was named in honor of Professor Helge Larsen, who pioneered research on halophiles.

Francisco Juan Martínez Mojica is a Spanish molecular biologist and microbiologist at the University of Alicante in Spain. He is known for his discovery of repetitive, functional DNA sequences in bacteria which he named CRISPR. These were later developed into the first widespread genome editing tool, CRISPR-Cas9.

Haloferacaceae is a family of halophilic, chemoorganotrophic or heterotrophic archaea within the order Haloferacales. The type genus of this family is Haloferax. Its biochemical characteristics are the same as the order Haloferacales.
Haloferax sulfurifontis is a species of archaea in the family Haloferacaceae.
Haloferax elongans is a species of archaea in the family Haloferacaceae.
Haloferax gibbonsii is a species of archaea in the family Haloferacaceae.

Haloferax mediterranei is a species of archaea in the family Haloferacaceae.
Haloferax mucosum is a species of archaea in the family Haloferacaceae.
Haloferax prahovense is a species of archaea in the family Haloferacaceae.
Haloferacales is an order of halophilic, chemoorganotrophic or heterotrophic archaea within the class Haloarchaea. The type genus of this order is Haloferax.






