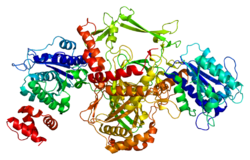
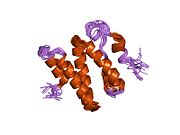
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.

Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.

Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.

Ku70 is a protein that, in humans, is encoded by the XRCC6 gene.

DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as PRKDC or XRCC7. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase consisting of a single polypeptide chain of 4,128 amino acids.

POU domain, class 2, transcription factor 1 is a protein that in humans is encoded by the POU2F1 gene.

Replication protein A 70 kDa DNA-binding subunit is a protein that in humans is encoded by the RPA1 gene.

DNA replication licensing factor MCM7 is a protein that in humans is encoded by the MCM7 gene.

Calcium/calmodulin-dependent protein kinase type II beta chain is an enzyme that in humans is encoded by the CAMK2B gene.

Cyclic AMP-dependent transcription factor ATF-1 is a protein that in humans is encoded by the ATF1 gene.

Calcium/calmodulin-dependent protein kinase type IV is an enzyme that in humans is encoded by the CAMK4 gene.

DNA replication licensing factor MCM4 is a protein that in humans is encoded by the MCM4 gene.

Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the KAT2A gene.

Eukaryotic translation initiation factor 2 subunit 2 (eIF2β) is a protein that in humans is encoded by the EIF2S2 gene.

A kinase anchor protein 1, mitochondrial is an enzyme that in humans is encoded by the AKAP1 gene.

Homeobox protein Hox-C4 is a protein that in humans is encoded by the HOXC4 gene.

A-kinase anchor protein 8 is an enzyme that, in humans, is encoded by the AKAP8 gene.
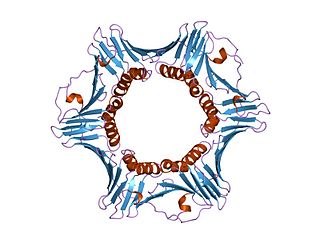
DNA polymerase delta subunit 3 is an enzyme that in humans is encoded by the POLD3 gene. It is a component of the DNA polymerase delta complex.

Interferon-alpha/beta receptor alpha chain is a protein that in humans is encoded by the IFNAR1 gene.