Related Research Articles
In developmental biology, photomorphogenesis is light-mediated development, where plant growth patterns respond to the light spectrum. This is a completely separate process from photosynthesis where light is used as a source of energy. Phytochromes, cryptochromes, and phototropins are photochromic sensory receptors that restrict the photomorphogenic effect of light to the UV-A, UV-B, blue, and red portions of the electromagnetic spectrum.
Shade avoidance is a set of responses that plants display when they are subjected to the shade of another plant. It often includes elongation, altered flowering time, increased apical dominance and altered partitioning of resources. This set of responses is collectively called the shade-avoidance syndrome (SAS).
Cryptochromes are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name cryptochrome was proposed as a portmanteau combining the chromatic nature of the photoreceptor, and the cryptogamic organisms on which many blue-light studies were carried out.
Florigens are proteins capable of inducing flowering time in Angiosperms. The prototypical florigen is encoded by the FT gene and its orthologs in Arabidopsis and other plants. Florigens are produced in the leaves, and act in the shoot apical meristem of buds and growing tips.
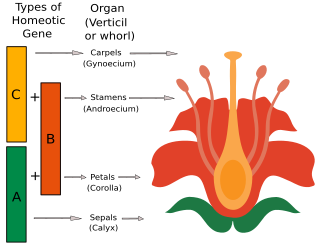
The ABC model of flower development is a scientific model of the process by which flowering plants produce a pattern of gene expression in meristems that leads to the appearance of an organ oriented towards sexual reproduction, a flower. There are three physiological developments that must occur in order for this to take place: firstly, the plant must pass from sexual immaturity into a sexually mature state ; secondly, the transformation of the apical meristem's function from a vegetative meristem into a floral meristem or inflorescence; and finally the growth of the flower's individual organs. The latter phase has been modelled using the ABC model, which aims to describe the biological basis of the process from the perspective of molecular and developmental genetics.
The repressilator is a genetic regulatory network consisting of at least one feedback loop with at least three genes, each expressing a protein that represses the next gene in the loop. In biological research, repressilators have been used to build cellular models and understand cell function. There are both artificial and naturally-occurring repressilators. Recently, the naturally-occurring repressilator clock gene circuit in Arabidopsis thaliana and mammalian systems have been studied.

In biology, phototropism is the growth of an organism in response to a light stimulus. Phototropism is most often observed in plants, but can also occur in other organisms such as fungi. The cells on the plant that are farthest from the light contain a hormone called auxin that reacts when phototropism occurs. This causes the plant to have elongated cells on the furthest side from the light. Phototropism is one of the many plant tropisms, or movements, which respond to external stimuli. Growth towards a light source is called positive phototropism, while growth away from light is called negative phototropism. Negative phototropism is not to be confused with skototropism, which is defined as the growth towards darkness, whereas negative phototropism can refer to either the growth away from a light source or towards the darkness. Most plant shoots exhibit positive phototropism, and rearrange their chloroplasts in the leaves to maximize photosynthetic energy and promote growth. Some vine shoot tips exhibit negative phototropism, which allows them to grow towards dark, solid objects and climb them. The combination of phototropism and gravitropism allow plants to grow in the correct direction.

Cycle (cyc) is a gene in Drosophila melanogaster that encodes the CYCLE protein (CYC). The Cycle gene (cyc) is expressed in a variety of cell types in a circadian manner. It is involved in controlling both the sleep-wake cycle and circadian regulation of gene expression by promoting transcription in a negative feedback mechanism. The cyc gene is located on the left arm of chromosome 3 and codes for a transcription factor containing a basic helix-loop-helix (bHLH) domain and a PAS domain. The 2.17 kb cyc gene is divided into 5 coding exons totaling 1,625 base pairs which code for 413 aminos acid residues. Currently 19 alleles are known for cyc. Orthologs performing the same function in other species include ARNTL and ARNTL2.
Timing of CAB expression 1 is a protein that in Arabidopsis thaliana is encoded by the TOC1 gene. TOC1 is also known as two-component response regulator-like APRR1.
Circadian Clock Associated 1 (CCA1) is a gene that is central to the circadian oscillator of angiosperms. It was first identified in Arabidopsis thaliana in 1993. CCA1 interacts with LHY and TOC1 to form the core of the oscillator system. CCA1 expression peaks at dawn. Loss of CCA1 function leads to a shortened period in the expression of many other genes.
Steve A. Kay is a British-born chronobiologist who mainly works in the United States. Dr. Kay has pioneered methods to monitor daily gene expression in real time and characterized circadian gene expression in plants, flies and mammals. In 2014, Steve Kay celebrated 25 years of successful chronobiology research at the Kaylab 25 Symposium, joined by over one hundred researchers with whom he had collaborated with or mentored. Dr. Kay, a member of the National Academy of Sciences, U.S.A., briefly served as president of The Scripps Research Institute. and is currently a professor at the University of Southern California. He also served on the Life Sciences jury for the Infosys Prize in 2011.
Andrew John McWalter Millar, FRS, FRSE is a Scottish chronobiologist, systems biologist, and molecular geneticist. Millar is a professor at The University of Edinburgh and also serves as its chair of systems biology. Millar is best known for his contributions to plant circadian biology; in the Steve Kay lab, he pioneered the use of luciferase imaging to identify circadian mutants in Arabidopsis. Additionally, Millar's group has implicated the ELF4 gene in circadian control of flowering time in Arabidopsis. Millar was elected to the Royal Society in 2012 and the Royal Society of Edinburgh in 2013.
Pseudo-response regulator (PRR) refers to a group of genes that regulate the circadian oscillator in plants. There are four primary PRR proteins that perform the majority of interactions with other proteins within the circadian oscillator, and another (PRR3) that has limited function. These genes are all paralogs of each other, and all repress the transcription of Circadian Clock Associated 1 (CCA1) and Late Elongated Hypocotyl (LHY) at various times throughout the day. The expression of PRR9, PRR7, PRR5 and TOC1/PRR1 peak around morning, mid-day, afternoon and evening, respectively. As a group, these genes are one part of the three-part repressilator system that governs the biological clock in plants.
The Late Elongated Hypocotyl gene (LHY), is an oscillating gene found in plants that functions as part of their circadian clock. LHY encodes components of mutually regulatory negative feedback loops with Circadian Clock Associated 1 (CCA1) in which overexpression of either results in dampening of both of their expression. This negative feedback loop affects the rhythmicity of multiple outputs creating a daytime protein complex. LHY was one of the first genes identified in the plant clock, along with TOC1 and CCA1. LHY and CCA1 have similar patterns of expression, which is capable of being induced by light. Single loss-of-function mutants in both genes result in seemingly identical phenotypes, but LHY cannot fully rescue the rhythm when CCA1 is absent, indicating that they may only be partially functionally redundant. Under constant light conditions, CCA1 and LHY double loss-of-function mutants fail to maintain rhythms in clock-controlled RNAs.
Transcription-translation feedback loop (TTFL) is a cellular model for explaining circadian rhythms in behavior and physiology. Widely conserved across species, the TTFL is auto-regulatory, in which transcription of clock genes is regulated by their own protein products.
Dmitri Nusinow is an American chronobiologist who studies plant circadian rhythms. He was born on November 7, 1976 in Inglewood, California. He currently resides in St. Louis, and his research focus includes a combination of molecular, biochemical, genetic, genomic, and proteomic tools to discover the molecular connections between signaling networks, circadian oscillators, and specific outputs. By combining these methods, he hopes to apply the knowledge elucidated from the Arabidopsis model to other plant species.
EARLY FLOWERING 3 (ELF3) is a plant-specific gene that encodes the hydroxyproline-rich glycoprotein and is required for the function of the circadian clock. ELF3 is one of the three components that make up the Evening Complex (EC) within the plant circadian clock, in which all three components reach peak gene expression and protein levels at dusk. ELF3 serves as a scaffold to bind EARLY FLOWERING 4 (ELF4) and LUX ARRHYTHMO (LUX), two other components of the EC, and functions to control photoperiod sensitivity in plants. ELF3 also plays an important role in temperature and light input within plants for circadian clock entrainment. Additionally, it plays roles in light and temperature signaling that are independent from its role in the EC.
Elaine Munsey Tobin is a professor of molecular, cell, and developmental biology at the University of California, Los Angeles (UCLA). Tobin is recognized as a Pioneer Member of the American Society of Plant Biologists (ASPB).
The chlorophyll a/b-binding protein gene, otherwise known as the CAB gene, is one of the most thoroughly characterized clock-regulated genes in plants. There are a variety of CAB proteins that are derived from this gene family. Studies on Arabidopsis plants have shed light on the mechanisms of biological clocks under the regulation of CAB genes. Dr. Steve Kay discovered that CAB was regulated by a circadian clock, which switched the gene on in the morning and off in the late afternoon. The genes code for proteins that associate with chlorophyll and xanthophylls. This association aids the absorption of sunlight, which transfers energy to photosystem II to drive photosynthetic electron transport.
Stacey Harmer is a chronobiologist whose work centers on the study of circadian rhythms in plants. Her research focuses on the molecular workings of the plant circadian clock and its influences on plant behaviors and physiology. She is a professor in the Department of Plant Biology at the University of California, Davis.
References
- 1 2 3 4 5 Troncoso-Ponce MA, Mas P (May 2012). "Newly described components and regulatory mechanisms of circadian clock function in Arabidopsis thaliana". Molecular Plant. 5 (3): 545–53. doi: 10.1093/mp/ssr117 . hdl: 10261/249493 . PMID 22230762.
- ↑ Yamashino T (2013-01-01). "From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana". Bioscience, Biotechnology, and Biochemistry. 77 (1): 10–6. doi:10.1271/bbb.120765. PMID 23291766. S2CID 33795797.
- ↑ Salanoubat M, Lemcke K, Rieger M, Ansorge W, Unseld M, Fartmann B, et al. (December 2000). "Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana". Nature. 408 (6814): 820–2. Bibcode:2000Natur.408..820E. doi:10.1038/35048706. PMID 11130713.
- ↑ Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, et al. (October 2003). "Empirical analysis of transcriptional activity in the Arabidopsis genome". Science. 302 (5646): 842–6. Bibcode:2003Sci...302..842Y. doi:10.1126/science.1088305. PMID 14593172. S2CID 7076927.
- ↑ Onai K, Ishiura M (October 2005). "PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock". Genes to Cells. 10 (10): 963–72. doi:10.1111/j.1365-2443.2005.00892.x. PMID 16164597. S2CID 40094451.
- ↑ "PCL1 Homeodomain-like superfamily protein [Arabidopsis thaliana (thale cress)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-04-14.
- 1 2 3 4 5 6 7 8 9 Huang H, Nusinow DA (October 2016). "Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock" (PDF). Trends in Genetics. 32 (10): 674–686. doi:10.1016/j.tig.2016.08.002. PMID 27594171.
- ↑ "LUX - Transcription factor LUX - Arabidopsis thaliana (Mouse-ear cress) - LUX gene & protein". www.uniprot.org. Retrieved 2017-04-14.
- 1 2 3 4 Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (July 2011). "The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth". Nature. 475 (7356): 398–402. doi:10.1038/nature10182. PMC 3155984 . PMID 21753751.
- 1 2 Liew LC, Hecht V, Sussmilch FC, Weller JL (June 2014). "The Pea Photoperiod Response Gene STERILE NODES Is an Ortholog of LUX ARRHYTHMO". Plant Physiology. 165 (2): 648–657. doi:10.1104/pp.114.237008. PMC 4044833 . PMID 24706549.
- 1 2 Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (September 2013). "HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways". The New Phytologist. 199 (4): 1045–59. doi:10.1111/nph.12346. PMC 3902989 . PMID 23731278.