A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
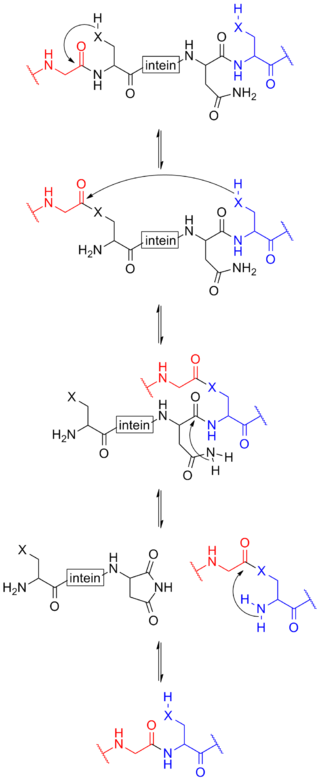
Protein splicing is an intramolecular reaction of a particular protein in which an internal protein segment is removed from a precursor protein with a ligation of C-terminal and N-terminal external proteins on both sides. The splicing junction of the precursor protein is mainly a cysteine or a serine, which are amino acids containing a nucleophilic side chain. The protein splicing reactions which are known now do not require exogenous cofactors or energy sources such as adenosine triphosphate (ATP) or guanosine triphosphate (GTP). Normally, splicing is associated only with pre-mRNA splicing. This precursor protein contains three segments—an N-extein followed by the intein followed by a C-extein. After splicing has taken place, the resulting protein contains the N-extein linked to the C-extein; this splicing product is also termed an extein.
Marlene Belfort is an American biochemist known for her research on the factors that interrupt genes and proteins. She is a fellow of the American Academy of Arts and Sciences and has been admitted to the United States National Academy of Sciences.
In molecular biology, a twintron is an intron-within-intron excised by sequential splicing reactions. A twintron is presumably formed by the insertion of a mobile intron into an existing intron.
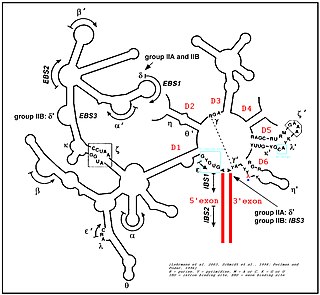
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and deliver cargo DNA such as reporter genes or lox sites

I-CreI is a homing endonuclease whose gene was first discovered in the chloroplast genome of Chlamydomonas reinhardtii, a species of unicellular green algae. It is named for the facts that: it resides in an Intron; it was isolated from Clamydomonas reinhardtii; it was the first (I) such gene isolated from C. reinhardtii. Its gene resides in a group I intron in the 23S ribosomal RNA gene of the C. reinhardtii chloroplast, and I-CreI is only expressed when its mRNA is spliced from the primary transcript of the 23S gene. I-CreI enzyme, which functions as a homodimer, recognizes a 22-nucleotide sequence of duplex DNA and cleaves one phosphodiester bond on each strand at specific positions. I-CreI is a member of the LAGLIDADG family of homing endonucleases, all of which have a conserved LAGLIDADG amino acid motif that contributes to their associative domains and active sites. When the I-CreI-containing intron encounters a 23S allele lacking the intron, I-CreI enzyme "homes" in on the "intron-minus" allele of 23S and effects its parent intron's insertion into the intron-minus allele. Introns with this behavior are called mobile introns. Because I-CreI provides for its own propagation while conferring no benefit on its host, it is an example of selfish DNA.

The homing endonucleases are a collection of endonucleases encoded either as freestanding genes within introns, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele frequency at greater than Mendelian rates.
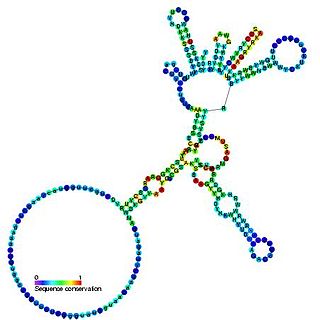
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

60S ribosomal protein L7a is a protein that in humans is encoded by the RPL7A gene.

40S ribosomal protein S3 is a protein that in humans is encoded by the RPS3 gene.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.
Meganucleases are endodeoxyribonucleases characterized by a large recognition site ; as a result this site generally occurs only once in any given genome. For example, the 18-base pair sequence recognized by the I-SceI meganuclease would on average require a genome twenty times the size of the human genome to be found once by chance. Meganucleases are therefore considered to be the most specific naturally occurring restriction enzymes.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
tRNA-intron lyase is an enzyme. As an endonuclease enzyme, tRNA-intron lyase is responsible for splicing phosphodiester bonds within non-coding ribonucleic acid chains. These non-coding RNA molecules form tRNA molecules after being processed, and this is dependent on tRNA-intron lyase to splice the pretRNA. tRNA processing is an important post-transcriptional modification necessary for tRNA maturation because it locates and removes introns in the pretRNA. This enzyme catalyses the following chemical reaction: