Cysteine is a semiessential proteinogenic amino acid with the formula HOOC−CH(−NH2)−CH2−SH. The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile.
In biochemistry, a disulfide refers to a functional group with the structure R−S−S−R′. The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. Persulfide usually refers to R−S−S−H compounds.

In organic chemistry, a thiol, or thiol derivative, is any organosulfur compound of the form R−SH, where R represents an alkyl or other organic substituent. The −SH functional group itself is referred to as either a thiol group or a sulfhydryl group, or a sulfanyl group. Thiols are the sulfur analogue of alcohols, and the word is a blend of "thio-" with "alcohol".

Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones.
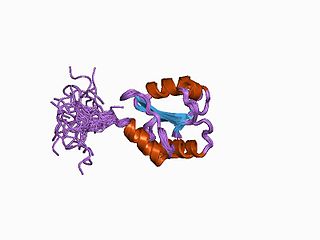
Protein disulfide isomerase, or PDI, is an enzyme in the endoplasmic reticulum (ER) in eukaryotes and the periplasm of bacteria that catalyzes the formation and breakage of disulfide bonds between cysteine residues within proteins as they fold. This allows proteins to quickly find the correct arrangement of disulfide bonds in their fully folded state, and therefore the enzyme acts to catalyze protein folding.
Native chemical ligation is an important extension of the chemical ligation concept for constructing a larger polypeptide chain by the covalent condensation of two or more unprotected peptides segments. Native chemical ligation is the most effective method for synthesizing native or modified proteins of typical size.
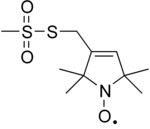
Site-directed spin labeling (SDSL) is a technique for investigating the structure and local dynamics of proteins using electron spin resonance. The theory of SDSL is based on the specific reaction of spin labels with amino acids. A spin label's built-in protein structure can be detected by EPR spectroscopy. SDSL is also a useful tool in examinations of the protein folding process.
CIDNP, often pronounced like "kidnip", is a nuclear magnetic resonance (NMR) technique that is used to study chemical reactions that involve radicals. It detects the non-Boltzmann (non-thermal) nuclear spin state distribution produced in these reactions as enhanced absorption or emission signals.

Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique to observe local magnetic fields around atomic nuclei. The sample is placed in a magnetic field and the NMR signal is produced by excitation of the nuclei sample with radio waves into nuclear magnetic resonance, which is detected with sensitive radio receivers. The intramolecular magnetic field around an atom in a molecule changes the resonance frequency, thus giving access to details of the electronic structure of a molecule and its individual functional groups. As the fields are unique or highly characteristic to individual compounds, in modern organic chemistry practice, NMR spectroscopy is the definitive method to identify monomolecular organic compounds.

Electron paramagnetic resonance (EPR) or electron spin resonance (ESR) spectroscopy is a method for studying materials that have unpaired electrons. The basic concepts of EPR are analogous to those of nuclear magnetic resonance (NMR), but the spins excited are those of the electrons instead of the atomic nuclei. EPR spectroscopy is particularly useful for studying metal complexes and organic radicals. EPR was first observed in Kazan State University by Soviet physicist Yevgeny Zavoisky in 1944, and was developed independently at the same time by Brebis Bleaney at the University of Oxford.
Nuclear magnetic resonance spectroscopy of proteins is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated.

2-Mercaptoethanol (also β-mercaptoethanol, BME, 2BME, 2-ME or β-met) is the chemical compound with the formula HOCH2CH2SH. ME or βME, as it is commonly abbreviated, is used to reduce disulfide bonds and can act as a biological antioxidant by scavenging hydroxyl radicals (amongst others). It is widely used because the hydroxyl group confers solubility in water and lowers the volatility. Due to its diminished vapor pressure, its odor, while unpleasant, is less objectionable than related thiols.

In chemistry, a sulfenic acid is an organosulfur compound and oxoacid with the general formula R−S−OH. It is the first member of the family of organosulfur oxoacids, which also include sulfinic acids and sulfonic acids, respectively. The base member of the sulfenic acid series with R = H is hydrogen thioperoxide.
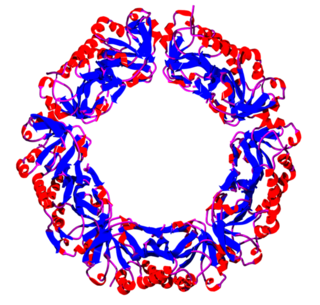
Peroxiredoxins are a ubiquitous family of antioxidant enzymes that also control cytokine-induced peroxide levels and thereby mediate signal transduction in mammalian cells. The family members in humans are PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6. The physiological importance of peroxiredoxins is indicated by their relative abundance. Their function is the reduction of peroxides, specifically hydrogen peroxide, alkyl hydroperoxides, and peroxynitrite.

TCEP is a reducing agent frequently used in biochemistry and molecular biology applications. It is often prepared and used as a hydrochloride salt (TCEP-HCl) with a molecular weight of 286.65 gram/mol. It is soluble in water and available as a stabilized solution at neutral pH and immobilized onto an agarose support to facilitate removal of the reducing agent.

Spin trapping is an analytical technique employed in chemistry and biology for the detection and identification of short-lived free radicals through the use of electron paramagnetic resonance (EPR) spectroscopy. EPR spectroscopy detects paramagnetic species such as the unpaired electrons of free radicals. However, when the half-life of radicals is too short to detect with EPR, compounds known as spin traps are used to react covalently with the radical products and form more stable adduct that will also have paramagnetic resonance spectra detectable by EPR spectroscopy. The use of radical-addition reactions to detect short-lived radicals was developed by several independent groups by 1968.

Chymopapain is a proteolytic enzyme isolated from the latex of papaya. It is a cysteine protease which belongs to the papain-like protease (PLCP) group. Because of its proteolytic activity, it is the main molecule in the process of chemonucleolysis, used in some procedures like the treatment of herniated lower lumbar discs in the spine by a nonsurgical method.

Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a strong constant magnetic field are perturbed by a weak oscillating magnetic field and respond by producing an electromagnetic signal with a frequency characteristic of the magnetic field at the nucleus. This process occurs near resonance, when the oscillation frequency matches the intrinsic frequency of the nuclei, which depends on the strength of the static magnetic field, the chemical environment, and the magnetic properties of the isotope involved; in practical applications with static magnetic fields up to ca. 20 tesla, the frequency is similar to VHF and UHF television broadcasts (60–1000 MHz). NMR results from specific magnetic properties of certain atomic nuclei. Nuclear magnetic resonance spectroscopy is widely used to determine the structure of organic molecules in solution and study molecular physics and crystals as well as non-crystalline materials. NMR is also routinely used in advanced medical imaging techniques, such as in magnetic resonance imaging (MRI).
Electron nuclear double resonance (ENDOR) is a magnetic resonance technique for elucidating the molecular and electronic structure of paramagnetic species. The technique was first introduced to resolve interactions in electron paramagnetic resonance (EPR) spectra. It is currently practiced in a variety of modalities, mainly in the areas of biophysics and heterogeneous catalysis.

Paramagnetic nuclear magnetic resonance spectroscopy refers to nuclear magnetic resonance (NMR) spectroscopy of paramagnetic compounds. Although most NMR measurements are conducted on diamagnetic compounds, paramagnetic samples are also amenable to analysis and give rise to special effects indicated by a wide chemical shift range and broadened signals. Paramagnetism diminishes the resolution of an NMR spectrum to the extent that coupling is rarely resolved. Nonetheless spectra of paramagnetic compounds provide insight into the bonding and structure of the sample. For example, the broadening of signals is compensated in part by the wide chemical shift range (often 200 ppm in 1H NMR). Since paramagnetism leads to shorter relaxation times (T1), the rate of spectral acquisition can be high.