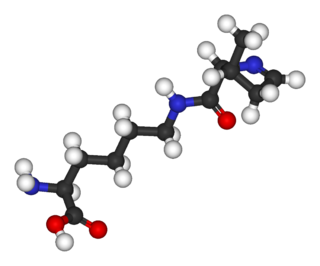
Pyrrolysine (symbol Pyl or O; encoded by the 'amber' stop codon UAG) is an ɑ-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group (which is in the protonated –+NH3 form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions). Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH.
A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:
Acidogenesis is the second stage in the four stages of anaerobic digestion:
Methanosarcina acetivorans is a versatile methane producing microbe which is found in such diverse environments as oil wells, trash dumps, deep-sea hydrothermal vents, and oxygen-depleted sediments beneath kelp beds. Only M. acetivorans and microbes in the genus Methanosarcina use all three known metabolic pathways for methanogenesis. Methanosarcinides, including M. acetivorans, are also the only archaea capable of forming multicellular colonies, and even show cellular differentiation. The genome of M. acetivorans is one of the largest archaeal genomes ever sequenced. Furthermore, one strain of M. acetivorans, M. a. C2A, has been identified to possess an F-type ATPase along with an A-type ATPase.

Methanosarcina is a genus of euryarchaeote archaea that produce methane. These single-celled organisms are known as anaerobic methanogens that produce methane using all three metabolic pathways for methanogenesis. They live in diverse environments where they can remain safe from the effects of oxygen, whether on the earth's surface, in groundwater, in deep sea vents, and in animal digestive tracts. Methanosarcina grow in colonies.
Methanococcus is a genus of coccoid methanogens of the family Methanococcaceae. They are all mesophiles, except the thermophilic M. thermolithotrophicus and the hyperthermophilic M. jannaschii. The latter was discovered at the base of a “white smoker” chimney at 21°N on the East Pacific Rise and it was the first archaeal genome to be completely sequenced, revealing many novel and eukaryote-like elements.

The 5,10-methenyltetrahydromethanopterin hydrogenase, the so-called iron-sulfur cluster-free hydrogenase, is an enzyme found in methanogenic archea such as Methanothermobacter marburgensis. It was discovered and first characterized by the Thauer group at the Max Planck Institute in Marburg. Hydrogenases are enzymes that either reduce protons or oxidize molecular dihydrogen.
In enzymology, a coenzyme F420 hydrogenase (EC 1.12.98.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a cytochrome-c3 hydrogenase (EC 1.12.2.1) is an enzyme that catalyzes the chemical reaction
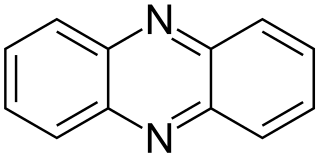
In enzymology, a Methanosarcina-phenazine hydrogenase (EC 1.12.98.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a CoB—CoM heterodisulfide reductase (EC 1.8.98.1) is an enzyme that catalyzes the chemical reaction
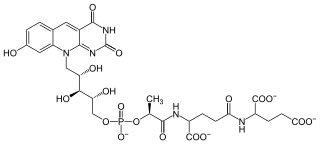
Coenzyme F420 or 8-hydroxy-5-deazaflavin is a coenzyme involved in redox reactions in methanogens, in many Actinobacteria, and sporadically in other bacterial lineages. It is a flavin derivative. The coenzyme is a substrate for coenzyme F420 hydrogenase, 5,10-methylenetetrahydromethanopterin reductase and methylenetetrahydromethanopterin dehydrogenase.

The flpD RNA motif is a conserved RNA structure discovered using bioinformatics. It is detected only in methanogenic archaea, and is generally located in the apparent 5' untranslated region of genes encoding hydrogenases that are likely involved in methane metabolism. For example, the flpD gene itself encodes a subunit of methyl-viologen-reducing hydrogenase.
Methanocaldococcus jannaschii is a thermophilic methanogenic archaean in the class Methanococci. It was the first archaeon to have its complete genome sequenced. The sequencing identified many genes unique to the archaea. Many of the synthesis pathways for methanogenic cofactors were worked out biochemically in this organism, as were several other archaeal-specific metabolic pathways.
The Methanosarcinales S-layer Tile Protein (MSTP) is a protein family found almost exclusively in Methanomicrobia members of the order Methanosarcinales. Typically a tandem repeat of two DUF1608 domains are contained in a single MSTP protein chain and these proteins self-assemble into the protective proteinaceous surface layer (S-layer) structure that encompasses the cell. The S-layer, which is found in most Archaea, and in many bacteria, serves many crucial functions including protection from deleterious extracellular substances.
Methanothrix soehngenii is a species of methanogenic archaea. Its cells are non-motile, non-spore-forming, rod-shaped and are normally combined end to end in long filaments, surrounded by a sheath-like structure. It is named in honour of N. L. Söhngen.
Methanolobus tindarius is a methanogen archaeon. It is marine, mesophilic, coccoid, lobal and monotrichous flagellated. They were isolated from coastal sediments.
Methanosarcina thermophila is a thermophilic, acetotrophic, methane-producing archaeon.
The H+-translocating F420H2 Dehydrogenase (F420H2DH) Family(TC# 3.D.9) is a member of the Na+ transporting Mrp superfamily. A single F420H2 dehydrogenase (also referred to as F420H2:quinol oxidoreductase) from the methanogenic archaeon, Methanosarcina mazei Gö1, has been shown to be a redox driven proton pump. The F420H2DH of M. mazei has a molecular size of about 120 kDa and contains Fe-S clusters and FAD. A similar five-subunit enzyme has been isolated from Methanolobus tindarius. The sulfate-reducing Archaeoglobus fulgidus (and several other archaea) also have this enzyme.