Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and can occur in the digestive tracts of animals including ruminants and humans, where they are responsible for the methane content of belching and flatulence. In marine sediments, the biological production of methane, termed methanogenesis, is generally confined to where sulfates are depleted below the top layers. Methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Other methanogens are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface.
A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:
Acidogenesis is the second stage in the four stages of anaerobic digestion:
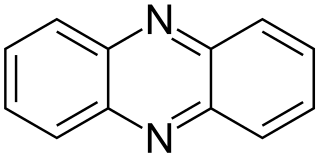
Phenazine is an organic compound with the formula (C6H4)2N2. It is a dibenzo annulated pyrazine, and the parent substance of many dyestuffs, such as the toluylene red, indulines, and safranines (and the closely related eurhodines). Phenazine crystallizes in yellow needles, which are only sparingly soluble in alcohol. Sulfuric acid dissolves it, forming a deep-red solution.
A hydrogenase mimic or bio-mimetic is an enzyme mimic of hydrogenases.

The 5,10-methenyltetrahydromethanopterin hydrogenase, the so-called iron-sulfur cluster-free hydrogenase, is an enzyme found in methanogenic archea such as Methanothermobacter marburgensis. It was discovered and first characterized by the Thauer group at the Max Planck Institute in Marburg. Hydrogenases are enzymes that either reduce protons or oxidize molecular dihydrogen.
In enzymology, a coenzyme F420 hydrogenase (EC 1.12.98.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a cytochrome-c3 hydrogenase (EC 1.12.2.1) is an enzyme that catalyzes the chemical reaction

In enzymology, ferredoxin hydrogenase, also referred to as [Fe-Fe]hydrogenase, H2 oxidizing hydrogenase, H2 producing hydrogenase, bidirectional hydrogenase, hydrogenase (ferredoxin), hydrogenlyase, and uptake hydrogenase, is found in Clostridium pasteurianum, Clostridium acetobutylicum,Chlamydomonas reinhardtii, and other organisms. The systematic name of this enzyme is hydrogen:ferredoxin oxidoreductase
In enzymology, a hydrogenase (acceptor) (EC 1.12.99.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogen dehydrogenase (EC 1.12.1.2) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogen dehydrogenase (NADP+) (EC 1.12.1.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogen:quinone oxidoreductase (EC 1.12.5.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a CoB—CoM heterodisulfide reductase (EC 1.8.98.1) is an enzyme that catalyzes the chemical reaction
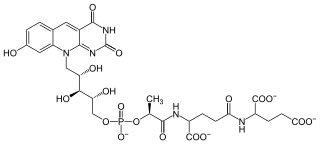
Coenzyme F420 or 8-hydroxy-5-deazaflavin is a coenzyme (sometimes called a cofactor) involved in redox reactions in methanogens, in many Actinomycetota, and sporadically in other bacterial lineages. It is a flavin derivative with an absorption maximum at 420 nm—hence its name. The coenzyme is a substrate for coenzyme F420 hydrogenase, 5,10-methylenetetrahydromethanopterin reductase and methylenetetrahydromethanopterin dehydrogenase.
Methanophenazine, a phenazine derivative, is a strongly hydrophobic redox-active cofactor with a role in electron transport in some methanogens. This chromophore can be purified from membranes of methanogenic archaea such as Methanosarcina mazei. The enzyme methanosarcina-phenazine hydrogenase has the name methanophenazine hydrogenase as a synonym.

Thermotoga maritima is a hyperthermophilic, anaerobic organism that is a member of the order Thermotogales. T. maritima is well known for its ability to produce hydrogen (clean energy) and it is the only fermentative bacterium that has been shown to produce Hydrogen more than the Thauer limit (>4 mol H2 /mol glucose). It employs [FeFe]-hydrogenases to produce hydrogen gas (H2) by fermenting many different types of carbohydrates.
[NiFe] hydrogenase is a type of hydrogenase, which is an oxidative enzyme that reversibly converts molecular hydrogen in prokaryotes including Bacteria and Archaea. The catalytic site on the enzyme provides simple hydrogen-metabolizing microorganisms a redox mechanism by which to store and utilize energy via the reaction
The H+-translocating F420H2 Dehydrogenase (F420H2DH) Family(TC# 3.D.9) is a member of the Na+ transporting Mrp superfamily. A single F420H2 dehydrogenase (also referred to as F420H2:quinol oxidoreductase) from the methanogenic archaeon, Methanosarcina mazei Gö1, has been shown to be a redox driven proton pump. The F420H2DH of M. mazei has a molecular size of about 120 kDa and contains Fe-S clusters and FAD. A similar five-subunit enzyme has been isolated from Methanolobus tindarius. The sulfate-reducing Archaeoglobus fulgidus (and several other archaea) also have this enzyme.
In enzymology, a formylmethanofuran dehydrogenase (EC 1.2.99.5) is an enzyme that catalyzes the chemical reaction:





