Methylation, in the chemical sciences, is the addition of a methyl group on a substrate, or the substitution of an atom by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and biology.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
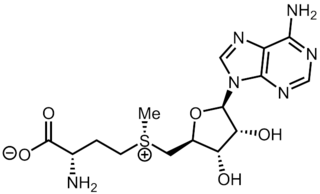
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
Histone-arginine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:histone-arginine Nomega-methyltransferase. This enzyme catalyses the following chemical reaction
In enzymology, a carnosine N-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a [cytochrome c]-lysine N-methyltransferase (EC 2.1.1.59) is an enzyme that catalyzes the chemical reaction

Enhancer of zeste homolog 2 (EZH2) is a histone-lysine N-methyltransferase enzyme encoded by EZH2 gene, that participates in histone methylation and, ultimately, transcriptional repression. EZH2 catalyzes the addition of methyl groups to histone H3 at lysine 27, by using the cofactor S-adenosyl-L-methionine. Methylation activity of EZH2 facilitates heterochromatin formation thereby silences gene function. Remodeling of chromosomal heterochromatin by EZH2 is also required during cell mitosis.

DOT1-like, histone H3K79 methyltransferase, also known as DOT1L, is a protein found in humans, as well as other eukaryotes.
Radical SAM enzymes belong to a superfamily of enzymes that use an iron-sulfur cluster (4Fe-4S) to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. Radical SAM enzymes comprise the largest superfamily of metal-containing enzymes.
Arsenite methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:arsenite As-methyltransferase. This enzyme catalyses the following chemical reaction
Glycine/sarcosine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:glycine(or sarcosine) N-methyltransferase . This enzyme catalyses the following chemical reaction
Sarcosine/dimethylglycine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:sarcosine(or N,N-dimethylglycine) N-methyltransferase . This enzyme catalyses the following chemical reaction
23S rRNA (uridine2552-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (uridine2552-2'-O-)-methyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (adenine2085-N6)-dimethyltransferase (EC 2.1.1.184, ErmC' methyltransferase, ermC methylase, ermC 23S rRNA methyltransferase, rRNA:m6A methyltransferase ErmC', ErmC', rRNA methyltransferase ErmC' ) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenine2085-N6)-dimethyltransferase. This enzyme catalyses the following chemical reaction
[Fructose-bisphosphate aldolase]-lysine N-methyltransferase is an enzyme that catalyses the following chemical reaction:
Protein methylation is a type of post-translational modification featuring the addition of methyl groups to proteins. It can occur on the nitrogen-containing side-chains of arginine and lysine, but also at the amino- and carboxy-termini of a number of different proteins. In biology, methyltransferases catalyze the methylation process, activated primarily by S-adenosylmethionine. Protein methylation has been most studied in histones, where the transfer of methyl groups from S-adenosyl methionine is catalyzed by histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.

Chuan He is a Chinese-American chemical biologist. He currently serves as the John T. Wilson Distinguished Service Professor at the University of Chicago, and an Investigator of the Howard Hughes Medical Institute. He is best known for his work in discovering and deciphering reversible RNA methylation in post-transcriptional gene expression regulation. He was awarded the 2023 Wolf Prize in Chemistry for his work in discovering and deciphering reversible RNA methylation in post-transcriptional gene expression regulation in addition to his contributions to the invention of TAB-seq, a biochemical method that can map 5-hydroxymethylcytosine (5hmC) at base-resolution genome-wide, as well as hmC-Seal, a method that covalently labels 5hmC for its detection and profiling.

Squire Booker is an American biochemist at The Pennsylvania State University. Booker directs an interdisciplinary chemistry research program related to fields of biochemistry, enzymology, protein chemistry, natural product biosynthesis, and mechanisms of radical dependent enzymes. He is an associate editor for the American Chemical Society Biochemistry Journal, is a Hughes Medical Institute Investigator, and an Eberly Distinguished Chair in Science at Penn State University.






