The cell is the basic structural and functional unit of all forms of life. Every cell consists of cytoplasm enclosed within a membrane; many cells contain organelles, each with a specific function. The term comes from the Latin word cellula meaning 'small room'. Most cells are only visible under a microscope. Cells emerged on Earth about 4 billion years ago. All cells are capable of replication, protein synthesis, and motility.
Cell biology is a branch of biology that studies the structure, function, and behavior of cells. All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and functioning of organisms. Cell biology is the study of the structural and functional units of cells. Cell biology encompasses both prokaryotic and eukaryotic cells and has many subtopics which may include the study of cell metabolism, cell communication, cell cycle, biochemistry, and cell composition. The study of cells is performed using several microscopy techniques, cell culture, and cell fractionation. These have allowed for and are currently being used for discoveries and research pertaining to how cells function, ultimately giving insight into understanding larger organisms. Knowing the components of cells and how cells work is fundamental to all biological sciences while also being essential for research in biomedical fields such as cancer, and other diseases. Research in cell biology is interconnected to other fields such as genetics, molecular genetics, molecular biology, medical microbiology, immunology, and cytochemistry.

Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there are two distinct types of cell division: a vegetative division (mitosis), producing daughter cells genetically identical to the parent cell, and a cell division that produces haploid gametes for sexual reproduction (meiosis), reducing the number of chromosomes from two of each type in the diploid parent cell to one of each type in the daughter cells. Mitosis is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is followed by telophase and cytokinesis; which divides the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the M phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells. To ensure proper progression through the cell cycle, DNA damage is detected and repaired at various checkpoints throughout the cycle. These checkpoints can halt progression through the cell cycle by inhibiting certain cyclin-CDK complexes. Meiosis undergoes two divisions resulting in four haploid daughter cells. Homologous chromosomes are separated in the first division of meiosis, such that each daughter cell has one copy of each chromosome. These chromosomes have already been replicated and have two sister chromatids which are then separated during the second division of meiosis. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
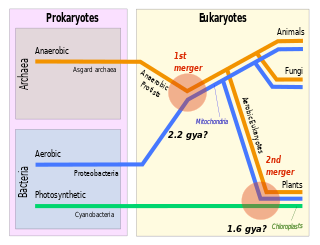
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.

Excavata is an extensive and diverse but paraphyletic group of unicellular Eukaryota. The group was first suggested by Simpson and Patterson in 1999 and the name latinized and assigned a rank by Thomas Cavalier-Smith in 2002. It contains a variety of free-living and symbiotic protists, and includes some important parasites of humans such as Giardia and Trichomonas. Excavates were formerly considered to be included in the now obsolete Protista kingdom. They were distinguished from other lineages based on electron-microscopic information about how the cells are arranged. They are considered to be a basal flagellate lineage.

Schizosaccharomyces pombe, also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs.
The diplomonads are a group of flagellates, most of which are parasitic. They include Giardia duodenalis, which causes giardiasis in humans. They are placed among the metamonads, and appear to be particularly close relatives of the retortamonads.

Karyogamy is the final step in the process of fusing together two haploid eukaryotic cells, and refers specifically to the fusion of the two nuclei. Before karyogamy, each haploid cell has one complete copy of the organism's genome. In order for karyogamy to occur, the cell membrane and cytoplasm of each cell must fuse with the other in a process known as plasmogamy. Once within the joined cell membrane, the nuclei are referred to as pronuclei. Once the cell membranes, cytoplasm, and pronuclei fuse, the resulting single cell is diploid, containing two copies of the genome. This diploid cell, called a zygote or zygospore can then enter meiosis, or continue to divide by mitosis. Mammalian fertilization uses a comparable process to combine haploid sperm and egg cells (gametes) to create a diploid fertilized egg.

Naegleria is a genus consisting of 47 described species of protozoa often found in warm aquatic environments as well as soil habitats worldwide. It has three life cycle forms: the amoeboid stage, the cyst stage, and the flagellated stage, and has been routinely studied for its ease in change from amoeboid to flagellated stages. The Naegleria genera became famous when Naegleria fowleri, the causative agent of the usually fatal human and animal disease primary amoebic meningoencephalitis (PAM), was discovered in 1965. Most species in the genus, however, are incapable of causing disease.
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. It adheres to Mendelian inheritance, with information coming from two parents, one male and one female—rather than matrilineally as in mitochondrial DNA.

Genome size is the total amount of DNA contained within one copy of a single complete genome. It is typically measured in terms of mass in picograms or less frequently in daltons, or as the total number of nucleotide base pairs, usually in megabases. One picogram is equal to 978 megabases. In diploid organisms, genome size is often used interchangeably with the term C-value.
Viral eukaryogenesis is the hypothesis that the cell nucleus of eukaryotic life forms evolved from a large DNA virus in a form of endosymbiosis within a methanogenic archaeon or a bacterium. The virus later evolved into the eukaryotic nucleus by acquiring genes from the host genome and eventually usurping its role. The hypothesis was first proposed by Philip Bell in 2001 and was further popularized with the discovery of large, complex DNA viruses that are capable of protein biosynthesis.

A nuclear gene is a gene that has its DNA nucleotide sequence physically situated within the cell nucleus of a eukaryotic organism. This term is employed to differentiate nuclear genes, which are located in the cell nucleus, from genes that are found in mitochondria or chloroplasts. The vast majority of genes in eukaryotes are nuclear.
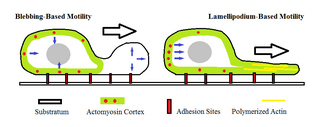
Amoeboid movement is the most typical mode of locomotion in adherent eukaryotic cells. It is a crawling-like type of movement accomplished by protrusion of cytoplasm of the cell involving the formation of pseudopodia ("false-feet") and posterior uropods. One or more pseudopodia may be produced at a time depending on the organism, but all amoeboid movement is characterized by the movement of organisms with an amorphous form that possess no set motility structures.
Willaertia /ˈwɪləɹʃə/ is a genus of non-pathogenic, free-living, thermophilic amoebae in the family Vahlkampfiidae.
The eukaryotes constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, and many unicellular organisms are eukaryotes. They constitute a major group of life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes.

Fission, in biology, is the division of a single entity into two or more parts and the regeneration of those parts to separate entities resembling the original. The object experiencing fission is usually a cell, but the term may also refer to how organisms, bodies, populations, or species split into discrete parts. The fission may be binary fission, in which a single organism produces two parts, or multiple fission, in which a single entity produces multiple parts.
Monocercomonoides is a genus of flagellate Excavata belonging to the order Oxymonadida. It was established by Bernard V. Travis and was first described as those with "polymastiginid flagellates having three anterior flagella and a trailing one originating at a single basal granule located in front of the anteriorly positioned nucleus, and a more or less well-defined axostyle". It is the first eukaryotic genus to be found to completely lack mitochondria, and all hallmark proteins responsible for mitochondrial function. The genus also lacks any other mitochondria related organelles (MROs) such as hydrogenosomes or mitosomes. Data suggests that the absence of mitochondria is not an ancestral feature, but rather due to secondary loss. Monocercomonoides sp. was found to obtain energy through an enzymatic action of nutrients absorbed from the environment. The genus has replaced the iron-sulfur cluster assembly pathway with a cytosolic sulfur mobilization system, likely acquired by horizontal gene transfer from a eubacterium of a common ancestor of oxymonads. These organisms are significant because they undermine assumptions that eukaryotes must have mitochondria to properly function. The genome of Monocercomonoides exilis has approximately 82 million base pairs, with 18 152 predicted protein-coding genes.
A linear chromosome is a chromosome which is linear in shape, and contains terminal ends. In most eukaryotic cells, DNA is arranged in multiple linear chromosomes. In contrast, most prokaryotic cells generally contain a singular circular chromosome.
The split gene theory is a theory of the origin of introns, long non-coding sequences in eukaryotic genes between the exons. The theory holds that the randomness of primordial DNA sequences would only permit small (< 600bp) open reading frames (ORFs), and that important intron structures and regulatory sequences are derived from stop codons. In this introns-first framework, the spliceosomal machinery and the nucleus evolved due to the necessity to join these ORFs into larger proteins, and that intronless bacterial genes are less ancestral than the split eukaryotic genes. The theory originated with Periannan Senapathy.