
An endospore is a dormant, tough, and non-reproductive structure produced by some bacteria in the phylum Bacillota. The name "endospore" is suggestive of a spore or seed-like form, but it is not a true spore. It is a stripped-down, dormant form to which the bacterium can reduce itself. Endospore formation is usually triggered by a lack of nutrients, and usually occurs in gram-positive bacteria. In endospore formation, the bacterium divides within its cell wall, and one side then engulfs the other. Endospores enable bacteria to lie dormant for extended periods, even centuries. There are many reports of spores remaining viable over 10,000 years, and revival of spores millions of years old has been claimed. There is one report of viable spores of Bacillus marismortui in salt crystals approximately 25 million years old. When the environment becomes more favorable, the endospore can reactivate itself into a vegetative state. Most types of bacteria cannot change to the endospore form. Examples of bacterial species that can form endospores include Bacillus cereus, Bacillus anthracis, Bacillus thuringiensis, Clostridium botulinum, and Clostridium tetani. Endospore formation is not found among Archaea.
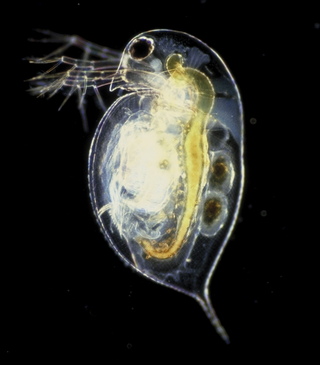
Daphnia is a genus of small planktonic crustaceans, 0.2–6.0 mm (0.01–0.24 in) in length. Daphnia are members of the order Anomopoda, and are one of the several small aquatic crustaceans commonly called water fleas because their saltatory swimming style resembles the movements of fleas. Daphnia spp. live in various aquatic environments ranging from acidic swamps to freshwater lakes and ponds.

Listeria monocytogenes is the species of pathogenic bacteria that causes the infection listeriosis. It is a facultative anaerobic bacterium, capable of surviving in the presence or absence of oxygen. It can grow and reproduce inside the host's cells and is one of the most virulent foodborne pathogens. Twenty to thirty percent of foodborne listeriosis infections in high-risk individuals may be fatal. In the European Union, listeriosis continues an upward trend that began in 2008, causing 2,161 confirmed cases and 210 reported deaths in 2014, 16% more than in 2013. In the EU, listeriosis mortality rates also are higher than those of other foodborne pathogens. Responsible for an estimated 1,600 illnesses and 260 deaths in the United States annually, listeriosis ranks third in total number of deaths among foodborne bacterial pathogens, with fatality rates exceeding even Salmonella spp. and Clostridium botulinum.
In evolutionary biology, an evolutionary arms race is an ongoing struggle between competing sets of co-evolving genes, phenotypic and behavioral traits that develop escalating adaptations and counter-adaptations against each other, resembling the geopolitical concept of an arms race. These are often described as examples of positive feedback. The co-evolving gene sets may be in different species, as in an evolutionary arms race between a predator species and its prey, or a parasite and its host. Alternatively, the arms race may be between members of the same species, as in the manipulation/sales resistance model of communication or as in runaway evolution or Red Queen effects. One example of an evolutionary arms race is in sexual conflict between the sexes, often described with the term Fisherian runaway. Thierry Lodé emphasized the role of such antagonistic interactions in evolution leading to character displacements and antagonistic coevolution.
The Red Queen's hypothesis is a hypothesis in evolutionary biology proposed in 1973, that species must constantly adapt, evolve, and proliferate in order to survive while pitted against ever-evolving opposing species. The hypothesis was intended to explain the constant (age-independent) extinction probability as observed in the paleontological record caused by co-evolution between competing species; however, it has also been suggested that the Red Queen hypothesis explains the advantage of sexual reproduction at the level of individuals, and the positive correlation between speciation and extinction rates in most higher taxa.

Thielaviopsis basicola is the plant-pathogen fungus responsible for black root rot disease. This particular disease has a large host range, affecting woody ornamentals, herbaceous ornamentals, agronomic crops, and even vegetable crops. Examples of susceptible hosts include petunia, pansy, poinsettia, tobacco, cotton, carrot, lettuce, tomato, and others. Symptoms of this disease resemble nutrient deficiency but are truly a result of the decaying root systems of plants. Common symptoms include chlorotic lower foliage, yellowing of plant, stunting or wilting, and black lesions along the roots. The lesions along the roots may appear red at first, getting darker and turning black as the disease progresses. Black root lesions that begin in the middle of a root can also spread further along the roots in either direction. Due to the nature of the pathogen, the disease can easily be identified by the black lesions along the roots, especially when compared to healthy roots. The black lesions that appear along the roots are a result of the formation of chlamydospores, resting spores of the fungus that contribute to its pathogenicity. The chlamydospores are a dark brown-black color and cause the "discoloration" of the roots when they are produced in large amounts.

Schistocephalus solidus is a tapeworm of fish, fish-eating birds and rodents. This hermaphroditic parasite belongs to the Eucestoda subclass, of class Cestoda. This species has been used to demonstrate that cross-fertilization produces a higher infective success rate than self-fertilization.

Bacillus anthracis is a gram-positive and rod-shaped bacterium that causes anthrax, a deadly disease to livestock and, occasionally, to humans. It is the only permanent (obligate) pathogen within the genus Bacillus. Its infection is a type of zoonosis, as it is transmitted from animals to humans. It was discovered by a German physician Robert Koch in 1876, and became the first bacterium to be experimentally shown as a pathogen. The discovery was also the first scientific evidence for the germ theory of diseases.
Dermocystidium is a genus of cyst-forming, eukaryotic fish parasites, the causative agents of dermocystidiosis.
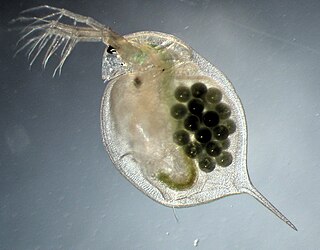
Daphnia magna is a small planktonic crustacean that belongs to the subclass Phyllopoda.
Host–parasite coevolution is a special case of coevolution, where a host and a parasite continually adapt to each other. This can create an evolutionary arms race between them. A more benign possibility is of an evolutionary trade-off between transmission and virulence in the parasite, as if it kills its host too quickly, the parasite will not be able to reproduce either. Another theory, the Red Queen hypothesis, proposes that since both host and parasite have to keep on evolving to keep up with each other, and since sexual reproduction continually creates new combinations of genes, parasitism favours sexual reproduction in the host.
Nosema bombi is a microsporidian, a small, unicellular parasite recently reclassified as a fungus that mainly affects bumble bees. It was reclassified as Vairimorpha bombi in 2020. The parasite infects numerous Bombus spp. at variable rates, and has been found to have a range of deleterious effects on its hosts.

The Pasteuriaceae are a family of nonmotile Gram-positive bacteria. They are moderately to strongly resistant to heat. Species in this family produce a septate mycelium with one refractile endospore. The mycelium grows bigger on one end to form sporangia and sometimes endospores. The size of the endospores is different for each species of the genus Pasteuria. Species of the family of Pastueriaceae are endoparasitic in plant, soil, and freshwater invertebrates.
Pasteuria is a genus of mycelial and endospore-forming, nonmotile gram-positive bacteria that are obligate parasites of some nematodes and crustaceans. The genus of Pasteuria was previously classified within the family Alicyclobacillaceae, but has since been moved to the family Pasteuriaceae.

Caenorhabditis elegans- microbe interactions are defined as any interaction that encompasses the association with microbes that temporarily or permanently live in or on the nematode C. elegans. The microbes can engage in a commensal, mutualistic or pathogenic interaction with the host. These include bacterial, viral, unicellular eukaryotic, and fungal interactions. In nature C. elegans harbours a diverse set of microbes. In contrast, C. elegans strains that are cultivated in laboratories for research purposes have lost the natural associated microbial communities and are commonly maintained on a single bacterial strain, Escherichia coli OP50. However, E. coli OP50 does not allow for reverse genetic screens because RNAi libraries have only been generated in strain HT115. This limits the ability to study bacterial effects on host phenotypes. The host microbe interactions of C. elegans are closely studied because of their orthologs in humans. Therefore, the better we understand the host interactions of C. elegans the better we can understand the host interactions within the human body.
Ordospora colligata is an intracellular parasite belonging to the Microsporidia. It is an obligatory gut parasite with the crustacean Daphnia magna as its only host. So far it has been reported from Europe and Asia.

Hamiltosporidium is a genus of Microsporidia, which are intracellular and unicellular parasites. The genus, proposed by Haag et al. in 2010, contains two species; Hamiltosporidium tvaerminnensis, and Hamiltosporidium magnivora. Both species infect only the crustacean Daphnia magna (Waterflea).

The study of gene-for-gene interactions uncovers genetic components, evolutionary impacts, and ecological/economic implications between rust fungi and plants. Rust fungi utilize the gene-for-gene interaction to invade host plants. Conversely, host plants utilize the gene-for-gene interaction to prevent invasion of rust fungi.

Dieter Ebert is professor for Zoology and Evolutionary Biology at the Zoological Institute at the University of Basel in Basel, Switzerland. He is an evolutionary ecologist and geneticist, known for his research on host–pathogen interaction and coevolution, mainly using the model system Daphnia and its parasites.

Phytoplankton are characterized as organisms which are unable to swim against a current and produce their own organic carbon via photosynthesis. They are responsible for producing approximately 50 percent of the Earth’s primary productivity and are therefore crucial in maintaining both marine ecosystems and adding a significant amount of oxygen to the atmosphere. However, as with other organisms, phytoplankton are hosts to many diverse forms of parasites, including, but not limited to, fungal- and non-fungal zoosporic parasites, Dinoflagellates, Cercozoans, and viruses. Parasites use nutrients from their hosts, at that organisms expense, and display diverse methods of infection. Parasites can play integral roles in the dynamics and interactions between phytoplankton and their communities, such as controlling population abundance, distribution and biodiversity.















