
Severe-acute-respiratory-syndrome–related coronavirus is a species of virus consisting of many known strains. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1, which caused the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing the ongoing pandemic of COVID-19. There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human mammal species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in Himalayan palm civets, which were likely ancestors of SARS-CoV-1.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the family Caulimoviridae, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.

Totiviridae is a family of double-stranded RNA viruses. Giardia lamblia, leishmania, trichomonas vaginalis, and fungi serve as natural hosts. The name of the group derives from Latin toti which means undivided or whole. There are 28 species in this family, assigned to 5 genera.

Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.

Nidovirales is an order of enveloped, positive-strand RNA viruses which infect vertebrates and invertebrates. Host organisms include mammals, birds, reptiles, amphibians, fish, arthropods, molluscs, and helminths. The order includes the families Coronaviridae, Arteriviridae, Roniviridae,Tobaniviridae, and Mesoniviridae.
Drosophila X virus (DXV) belongs to the Birnaviridae family of viruses. Birnaviridae currently consists of three genera. The first genus is Entomobirnavirus, which contains DXV. The next genus is Aquabirnavirus, containing infectious pancreatic necrosis virus (IPNV). The last genus is Avibirnavirus, which contains infectious bursal disease virus (IBDV). All of these genera contain homology in three specific areas of their transcripts. The homology comes from the amino and carboxyl regions of preVP2, a small 21-residue-long domain near the carboxyl terminal of VP3, and similar small ORFs sequences.

Deformed wing virus (DWV) is an RNA virus, one of 22 known viruses affecting honey bees. While most commonly infecting the honey bee, Apis mellifera, it has also been documented in other bee species, like Bombus terrestris, thus, indicating it may have a wider host specificity than previously anticipated. The virus was first isolated from a sample of symptomatic honeybees from Japan in the early 1980s and is currently distributed worldwide. It is found also in pollen baskets and commercially reared bumblebees. Its main vector in A. mellifera is the Varroa mite. It is named after what is usually the most obvious deformity it induces in the development of a honeybee pupa, which is shrunken and deformed wings, but other developmental deformities are often present.

HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence.

The 3C-like protease (3CLpro) or main protease (Mpro), formally known as C30 endopeptidase or 3-chymotrypsin-like protease, is the main protease found in coronaviruses. It cleaves the coronavirus polyprotein at eleven conserved sites. It is a cysteine protease and a member of the PA clan of proteases. It has a cysteine-histidine catalytic dyad at its active site and cleaves a Gln–(Ser/Ala/Gly) peptide bond.
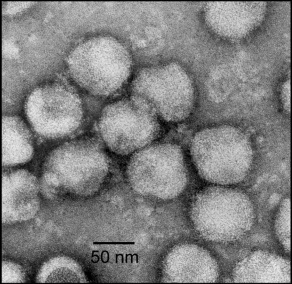
Mesoniviridae is a family of enveloped, positive-strand RNA viruses in the order Nidovirales which infect mosquitoes. The family is named after the size of the genomes relative to other nidoviruses, with meso- coming from the Greek word mesos, which means medium, and -ni being an abbreviation of nido.
Entebbe bat virus is an infectious disease caused by a Flavivirus that is closely related to yellow fever.

Positive-strand RNA viruses are a group of related viruses that have positive-sense, single-stranded genomes made of ribonucleic acid. The positive-sense genome can act as messenger RNA (mRNA) and can be directly translated into viral proteins by the host cell's ribosomes. Positive-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) which is used during replication of the genome to synthesize a negative-sense antigenome that is then used as a template to create a new positive-sense viral genome.
Alphamononivirus is a genus of enveloped, positive-strand RNA viruses in the order Nidovirales which infect planarian flatworms. Member virus planarian secretory cell nidovirus (PSCNV) has the largest known nonsegmented RNA genome of 41.1kb of any RNA virus. The genus is monotypic. It contains the subgenus Dumedivirus, which contains only one species, Planidovirus 1. Alphamononivirus is also the only member of the subfamily Mononivirinae, which in turn is the only member of family Mononiviridae, which likewise is the only member of the Monidovirineae suborder.

Riboviria is a realm of viruses that includes all viruses that use a homologous RNA-dependent polymerase for replication. It includes RNA viruses that encode an RNA-dependent RNA polymerase, as well as reverse-transcribing viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.

Orthornavirae is a kingdom of viruses that have genomes made of ribonucleic acid (RNA), including genes which encode an RNA-dependent RNA polymerase (RdRp). The RdRp is used to transcribe the viral RNA genome into messenger RNA (mRNA) and to replicate the genome. Viruses in this kingdom share a number of characteristics which promote rapid evolution, including high rates of genetic mutation, recombination, and reassortment.
Rio Negro virus is an alphavirus that was first isolated in Argentina in 1980. The virus was first called Ag80-663 but was renamed to Rio Negro virus in 2005. It is a former member of the Venezuelan equine encephalitis complex (VEEC), which are a group of alphaviruses in the Americas that have the potential to emerge and cause disease. Río Negro virus was recently reclassified as a distinct species. Closely related viruses include Mucambo virus and Everglades virus.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.

The nidoviral papain-like protease is a papain-like protease protein domain encoded in the genomes of nidoviruses. It is expressed as part of a large polyprotein from the ORF1a gene and has cysteine protease enzymatic activity responsible for proteolytic cleavage of some of the N-terminal viral nonstructural proteins within the polyprotein. A second protease also encoded by ORF1a, called the 3C-like protease or main protease, is responsible for the majority of further cleavages. Coronaviruses have one or two papain-like protease domains; in SARS-CoV and SARS-CoV-2, one PLPro domain is located in coronavirus nonstructural protein 3 (nsp3). Arteriviruses have two to three PLP domains. In addition to their protease activity, PLP domains function as deubiquitinating enzymes (DUBs) that can cleave the isopeptide bond found in ubiquitin chains. They are also "deISGylating" enzymes that remove the ubiquitin-like domain interferon-stimulated gene 15 (ISG15) from cellular proteins. These activities are likely responsible for antagonizing the activity of the host innate immune system. Because they are essential for viral replication, papain-like protease domains are considered drug targets for the development of antiviral drugs against human pathogens such as MERS-CoV, SARS-CoV, and SARS-CoV-2.

Nsp12 is a non-structural protein in the Coronavirus genome. Its gene is part of the ORF1ab reading frame and it is part of the pp1ab polyprotein; it is cleaved by 3CLpro.














