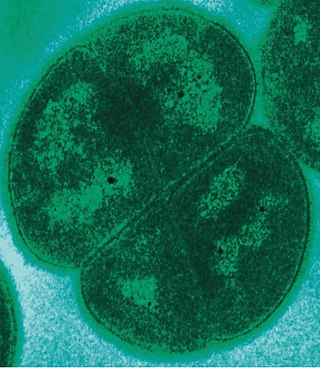
Deinococcota is a phylum of bacteria with a single class, Deinococci, that are highly resistant to environmental hazards, also known as extremophiles. These bacteria have thick cell walls that give them gram-positive stains, but they include a second membrane and so are closer in structure to those of gram-negative bacteria.

"Candidatus Pelagibacter", with the single species "Ca. P. communis", was isolated in 2002 and given a specific name, although it has not yet been described as required by the bacteriological code. It is an abundant member of the SAR11 clade in the phylum Alphaproteobacteria. SAR11 members are highly dominant organisms found in both salt and fresh water worldwide and were originally known only from their rRNA genes, first identified in the Sargasso Sea in 1990 by Stephen Giovannoni's laboratory at Oregon State University and later found in oceans worldwide. "Ca. P. communis" and its relatives may be the most abundant organisms in the ocean, and quite possibly the most abundant bacteria in the entire world. It can make up about 25% of all microbial plankton cells, and in the summer they may account for approximately half the cells present in temperate ocean surface water. The total abundance of "Ca. P. communis" and relatives is estimated to be about 2 × 1028 microbes.
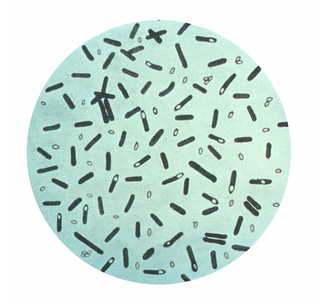
The Clostridiaceae are a family of the bacterial class Clostridia, and contain the genus Clostridium.
Alteromonas is a genus of Pseudomonadota found in sea water, either in the open ocean or in the coast. It is Gram-negative. Its cells are curved rods with a single polar flagellum.
Pseudoalteromonas piscicida is a marine bacterium. It is known to produce a quorum sensing molecule called 2-heptyl-4-quinolone (HHQ), which functions as a bacterial infochemical. Research into the effects of this infochemical on phytoplankton is currently being conducted by Dr. Kristen Whalen of Haverford College.
In taxonomy, Methanothermococcus is a genus of the Methanococcaceae. The cells are shaped like irregular bars and tend to be Gram-negative. They are mobile via polar flagella. They require acetate to grow.

60S ribosomal protein L13a is a protein that in humans is encoded by the RPL13A gene.

60S ribosomal protein L35a is a protein that in humans is encoded by the RPL35A gene.

60S ribosomal protein L27a is a protein that in humans is encoded by the RPL27A gene.
Algicola is a genus in the phylum Pseudomonadota (Bacteria).

'The All-Species Living Tree' Project is a collaboration between various academic groups/institutes, such as ARB, SILVA rRNA database project, and LPSN, with the aim of assembling a database of 16S rRNA sequences of all validly published species of Bacteria and Archaea. At one stage, 23S sequences were also collected, but this has since stopped.
Pseudoalteromonas aurantia is an antibacterial-producing marine bacterium commonly found in Mediterranean waters. In 1979, Gauthier and Breittmayer first named it Alteromonas aurantia to include it in the genus Alteromonas that was described seven years earlier, in 1972 by Baumann et al. In 1995, Gauthier et al renamed Alteromonas aurantia to Pseudoalteromonas aurantia to include it in their proposed new genus, Pseudoalteromonas, which they recommended splitting from Alteromonas.
Pseudoalteromonas citrea is a yellow-pigmented marine bacterium that is antibiotic-producing and was isolated from Mediterranean waters off Nice. Originally named Alteromonas citrea, nearly two decades later it was reclassified as part of the Genus Pseudoalteromonas.
Pseudoalteromonas distincta is a marine bacterium.
Pseudoalteromonas espejiana is a marine bacterium.
Pseudoalteromonas luteoviolacea is a marine bacterium which was isolated from seawater near Nice.
Pseudoalteromonas rubra is a marine bacterium.
Pseudoalteromonas tetraodonis is a marine bacterium isolated from the surface slime of the puffer fish. It secretes the neurotoxin, tetrodotoxin. It was originally described in 1990 as Alteromonas tetraodonis but was reclassified in 2001 to the genus Pseudoalteromonas.
Pseudoalteromonas undina is a marine bacterium isolated from seawater off the coast of Northern California. It was originally classified as Alteromonas undina but was reclassified in 1995 to the genus Pseudoalteromonas.
Alteromonas macleodii is a species of widespread marine bacterium found in surface waters across temperate and tropical regions. First discovered in a survey of aerobic bacteria in 1972, A. macleodii has since been placed within the phylum Pseudomonadota and is recognised as a prominent component of surface waters between 0 and 50 metres. Alteromonas macleodii has a single circular DNA chromosome of 4.6 million base pairs. Variable regions in the genome of A. macleodii confer functional diversity to closely related strains and facilitate different lifestyles and strategies. Certain A. macleodii strains are currently being explored for their industrial uses, including in cosmetics, bioethanol production and rare earth mining.






