
Pseudomonas is a genus of Gram-negative bacteria belonging to the family Pseudomonadaceae in the class Gammaproteobacteria. The 313 members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.

Pseudomonas fluorescens is a common Gram-negative, rod-shaped bacterium. It belongs to the Pseudomonas genus; 16S rRNA analysis as well as phylogenomic analysis has placed P. fluorescens in the P. fluorescens group within the genus, to which it lends its name.

Pseudomonas putida is a Gram-negative, rod-shaped, saprophytic soil bacterium. It has a versatile metabolism and is amenable to genetic manipulation, making it a common organism used in research, bioremediation, and synthesis of chemicals and other compounds.
Pseudomonas chlororaphis is a bacterium used as a soil inoculant in agriculture and horticulture. It can act as a biocontrol agent against certain fungal plant pathogens via production of phenazine-type antibiotics. Based on 16S rRNA analysis, similar species have been placed in its group.
Pseudomonas fragi is a psychrophilic, Gram-negative bacterium that is responsible for dairy spoilage. Unlike many other members of the genus Pseudomonas, P. fragi does not produce siderophores. Optimal temperature for growth is 30 °C, however it can grow between 0 and 35 °C. Based on 16S rRNA analysis, P. fragi has been placed in the P. chlororaphis group.
Pseudomonas corrugata is a Gram-negative, plant-pathogenic bacterium that causes pith necrosis in tomatoes. Based on 16S rRNA analysis, P. corrugata has been placed in the P. fluorescens group. For a more comprehensive phylogenetic analysis of P. corrugata and its closely related phytopathogenic bacterium P. mediterranea, refer to Trantas et al. 2015.
Pseudomonas citronellolis is a Gram-negative, bacillus bacterium that is used to study the mechanisms of pyruvate carboxylase. It was first isolated from forest soil, under pine trees, in northern Virginia, United States.
Pseudomonas denitrificans is a Gram-negative aerobic bacterium that performs denitrification. It was first isolated from garden soil in Vienna, Austria. It overproduces cobalamin (vitamin B12), which it uses for methionine synthesis and it has been used for manufacture of the vitamin. Scientists at Rhône-Poulenc Rorer took a genetically engineered strain of the bacteria, in which eight of the cob genes involved in the biosynthesis of the vitamin had been overexpressed, to establish the complete sequence of methylation and other steps in the cobalamin pathway.
Pseudomonas oleovorans is a Gram-negative, methylotrophic bacterium that is a source of rubredoxin. It was first isolated in water-oil emulsions used as lubricants and cooling agents for cutting metals. Based on 16S rRNA analysis, P. oleovorans has been placed in the P. aeruginosa group.
Pseudomonas lundensis is a Gram-negative, rod-shaped bacterium that often causes spoilage of milk, cheese, meat, and fish. Based on 16S rRNA analysis, P. lundensis has been placed in the P. chlororaphis group.
Pseudomonas taetrolens is a Gram-negative, nonsporulating, motile, rod-shaped bacterium that causes mustiness in eggs. Based on 16S rRNA analysis, P. taetrolens has been placed in the P. chlororaphis group.
Pseudomonas cedrina is a Gram-negative, rod-shaped bacterium isolated from spring waters in Lebanon. Based on 16S rRNA analysis, P. cedrina has been placed in the P. fluorescens group.
Pseudomonas orientalis is a Gram-negative, rod-shaped bacterium isolated from spring waters in Lebanon. Based on 16S rRNA analysis, P. orientalis has been placed in the P. fluorescens group.
Pseudomonas mandelii is a fluorescent, Gram-negative, rod-shaped bacterium isolated from natural spring waters in France. Based on 16S rRNA analysis, P. mandelii has been placed in the P. fluorescens group.
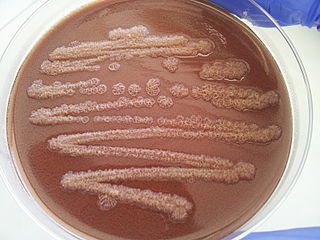
Pseudomonas stutzeri is a Gram-negative soil bacterium that is motile, has a single polar flagellum, and is classified as bacillus, or rod-shaped. While this bacterium was first isolated from human spinal fluid, it has since been found in many different environments due to its various characteristics and metabolic capabilities. P. stutzeri is an opportunistic pathogen in clinical settings, although infections are rare. Based on 16S rRNA analysis, this bacterium has been placed in the P. stutzeri group, to which it lends its name.
Pseudomonas nitroreducens is an aerobic, Gram-negative soil bacterium first isolated from oil brine in Japan. It is able to synthesise polyhydroxybutyrate homopolymer from medium chain length fatty acids. Based on 16S rRNA analysis, P. nitroreducens has been placed in the P. aeruginosa group.
'Pseudomonas' boreopolis is a species of Gram-negative bacteria. Following 16S rRNA phylogenetic analysis, it was determined that 'P.' boreopolis belonged in the Xanthomonas—Xylella rRNA lineage. It has not yet been further classified.
'Pseudomonas' carboxydohydrogena is a species of Gram-negative bacteria. Following 16S rRNA phylogenetic analysis, it was determined that 'P.' carboxydohydrogena belonged in the Bradyrhizobium rRNA lineage. It has not yet been further classified.

2,4-Diacetylphloroglucinol or Phl is a natural phenol found in several bacteria:
Pseudomonas protegens are widespread Gram-negative, plant-protecting bacteria. Some of the strains of this novel bacterial species previously belonged to P. fluorescens. They were reclassified since they seem to cluster separately from other fluorescent Pseudomonas species. P. protegens is phylogenetically related to the Pseudomonas species complexes P. fluorescens, P. chlororaphis, and P. syringae. The bacterial species characteristically produces the antimicrobial compounds pyoluteorin and 2,4-diacetylphloroglucinol (DAPG) which are active against various plant pathogens.




