
Pseudomonas is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.

Pseudomonas fluorescens is a common Gram-negative, rod-shaped bacterium. It belongs to the Pseudomonas genus; 16S rRNA analysis as well as phylogenomic analysis has placed P. fluorescens in the P. fluorescens group within the genus, to which it lends its name.

Acinetobacter is a genus of gram-negative bacteria belonging to the wider class of Gammaproteobacteria. Acinetobacter species are oxidase-negative, exhibit twitching motility, and occur in pairs under magnification.

Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants, humans and marine sponges. As a member of the genus Bacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.

Pseudomonas aeruginosa is a common encapsulated, gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.

Antibiotic sensitivity testing or antibiotic susceptibility testing is the measurement of the susceptibility of bacteria to antibiotics. It is used because bacteria may have resistance to some antibiotics. Sensitivity testing results can allow a clinician to change the choice of antibiotics from empiric therapy, which is when an antibiotic is selected based on clinical suspicion about the site of an infection and common causative bacteria, to directed therapy, in which the choice of antibiotic is based on knowledge of the organism and its sensitivities.

Shigella flexneri is a species of Gram-negative bacteria in the genus Shigella that can cause diarrhea in humans. Several different serogroups of Shigella are described; S. flexneri belongs to group B. S. flexneri infections can usually be treated with antibiotics, although some strains have become resistant. Less severe cases are not usually treated because they become more resistant in the future. Shigella are closely related to Escherichia coli, but can be differentiated from E.coli based on pathogenicity, physiology and serology.
Thermus thermophilus is a Gram-negative bacterium used in a range of biotechnological applications, including as a model organism for genetic manipulation, structural genomics, and systems biology. The bacterium is extremely thermophilic, with an optimal growth temperature of about 65 °C (149 °F). Thermus thermophilus was originally isolated from a thermal vent within a hot spring in Izu, Japan by Tairo Oshima and Kazutomo Imahori. The organism has also been found to be important in the degradation of organic materials in the thermogenic phase of composting. T. thermophilus is classified into several strains, of which HB8 and HB27 are the most commonly used in laboratory environments. Genome analyses of these strains were independently completed in 2004.
Pseudomonas chlororaphis is a bacterium used as a soil inoculant in agriculture and horticulture. It can act as a biocontrol agent against certain fungal plant pathogens via production of phenazine-type antibiotics. Based on 16S rRNA analysis, similar species have been placed in its group.
Burkholderia gladioli is a species of aerobic gram-negative rod-shaped bacteria that causes disease in both humans and plants. It can also live in symbiosis with plants and fungi and is found in soil, water, the rhizosphere, and in many animals. It was formerly known as Pseudomonas marginata.
Pseudomonas veronii is a Gram-negative, rod-shaped, fluorescent, motile bacterium isolated from natural springs in France. It may be used for bioremediation of contaminated soils, as it has been shown to degrade a variety of simple aromatic organic compounds. Based on 16S rRNA analysis, P. veronii has been placed in the P. fluorescens group.
Pseudomonas balearica is a Gram-negative, rod-shaped, nonfluorescent, motile, and denitrifying bacterium. It is an environmental bacterium that has been mostly isolated from polluted environments all over the world. Many of the isolates have demonstrated capabilities to degrade several compounds. Some of the strains are naphthalene degraders and one strain isolated in New Zealand has demonstrated the potential to oxidize inorganic sulfur compounds to tetrathionate. Based on 16S rRNA analysis, P. balearica has been placed in the P. stutzeri group.
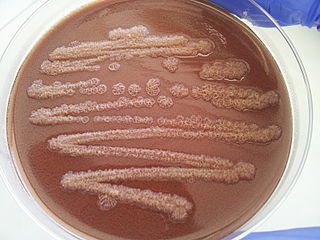
Pseudomonas stutzeri is a Gram-negative soil bacterium that is motile, has a single polar flagellum, and is classified as bacillus, or rod-shaped. While this bacterium was first isolated from human spinal fluid, it has since been found in many different environments due to its various characteristics and metabolic capabilities. P. stutzeri is an opportunistic pathogen in clinical settings, although infections are rare. Based on 16S rRNA analysis, this bacterium has been placed in the P. stutzeri group, to which it lends its name.
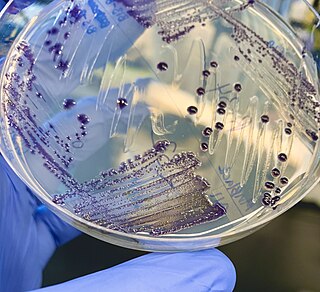
The genus Massilia belongs to the family Oxalobacteriaceae, and describes a group of gram-negative, motile, rod-shaped cells. They may contain either peritrichous or polar flagella. This genus was first described in 1998, after the type species, Massilia timonae, was isolated from the blood of an immunocompromised patient. The genus was named after the old Greek and Roman name for the city of Marseille, France, where the organism was first isolated. The Massilia genus is a diverse group that resides in many different environments, has many heterotrophic means of gathering energy, and is commonly found in association with plants.

Latilactobacillus sakei is the type species of the genus Latilactobacillus that was previously classified in the genus Lactobacillus. It is homofermentative; hexoses are metabolized via glycolysis to lactic acid as main metabolite; pentoses are fermented via the Phosphoketolase pathway to lactic and acetic acids.
Parachlamydia acanthamoebae are bacterium that fall into the category of host-associated microorganisms. This bacterium lives within free-living amoebae that are an intricate part of their reproduction. Originally named Candidatus Parachlamydia acanthamoebae, its current scientific name was introduced shortly after. This species has shown to have over eighty percent 16S rRNA gene sequencing identity with the class Chlamydiia. Parachlamydia acanthamoebae has the same family as the genus Neochlamydia with which it shares many similarities.
Alcanivorax pacificus is a pyrene-degrading marine gammaproteobacterium. It is of the genus Alcanivorax, a group of marine bacteria known for degrading hydrocarbons. When originally proposed, the genus Alcanivorax comprised six distinguishable species. However, A. pacificus, a seventh strain, was isolated from deep sea sediments in the West Pacific Ocean by Shanghai Majorbio Bio-pharm Technology Co., Ltd. in 2011. A. pacificus’s ability to degrade hydrocarbons can be employed for cleaning up oil-contaminated oceans through bioremediation. The genomic differences present in this strain of Alcanivorax that distinguish it from the original consortium are important to understand to better utilize this bacteria for bioremediation.
Dehalogenimonas lykanthroporepellens is an anaerobic, Gram-negative bacteria in the phylum Chloroflexota isolated from a Superfund site in Baton Rouge, Louisiana. It is useful in bioremediation for its ability to reductively dehalogenate chlorinated alkanes.
Xenophilus azovorans is a bacterium from the genus Xenophilus which has been isolated from soil in Switzerland.
Helicobacter typhlonius is a Gram-negative bacterium and opportunistic pathogen found in the genus Helicobacter. Only 35 known species are in this genus, which was described in 1982. H. typhlonius has a small number of close relatives, including Helicobacter muridarum, Helicobacter trogontum, and Helicobacter hepaticus, with the latter being the closest relative and much more prevalent.









