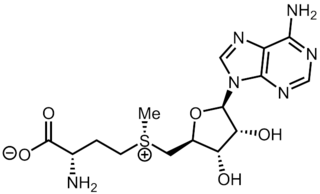
S-Adenosyl methionine (SAM-e) is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM-e is produced and consumed in the liver. More than 40 methyl transfers from SAM-e are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM-e was first discovered by Giulio Cantoni in 1952.

Methionine synthase also known as MS, MeSe, MetH is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the MTR gene. Methionine synthase forms part of the S-adenosylmethionine (SAMe) biosynthesis and regeneration cycle. In animals this enzyme requires Vitamin B12 (cobalamin) as a cofactor, whereas the form found in plants is cobalamin-independent. Microorganisms express both cobalamin-dependent and cobalamin-independent forms.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltrasferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the nucleophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

S-Adenosyl-l-homocysteine (SAH) is the biosynthetic precursor to homocysteine. SAH is formed by the demethylation of S-adenosyl-l-methionine. Adenosylhomocysteinase converts SAH into homocysteine and adenosine.

The PDE2 enzyme is one of 21 different phosphodiesterases (PDE) found in mammals. These different PDEs can be subdivided to 11 families. The different PDEs of the same family are functionally related despite the fact that their amino acid sequences show considerable divergence. The PDEs have different substrate specificities. Some are cAMP selective hydrolases, others are cGMP selective hydrolases and the rest can hydrolyse both cAMP and cGMP.
Protein L-isoaspartyl methyltransferase , also called S-adenosyl-L-methionine:protein-L-isoaspartate O-methyltransferase, is an enzyme which recognizes and catalyzes the repair of damaged L-isoaspartyl and D-aspartyl groups in proteins. It is a highly conserved enzyme which is present in nearly all eukaryotes, archaebacteria, and Gram-negative eubacteria.

Cystathionine-β-synthase, also known as CBS, is an enzyme (EC 4.2.1.22) that in humans is encoded by the CBS gene. It catalyzes the first step of the transsulfuration pathway, from homocysteine to cystathionine:

Adenosylhomocysteinase (EC 3.3.1.1, S-adenosylhomocysteine synthase, S-adenosylhomocysteine hydrolase, adenosylhomocysteine hydrolase, S-adenosylhomocysteinase, SAHase, AdoHcyase) is an enzyme that converts S-adenosylhomocysteine to homocysteine and adenosine. This enzyme catalyses the following chemical reaction

Guanidinoacetate N-methyltransferase is an enzyme that catalyzes the chemical reaction and is encoded by gene GAMT located on chromosome 19p13.3.
In enzymology, a jasmonate O-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a mRNA (guanine-N7-)-methyltransferase also known as mRNA cap guanine-N7 methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a mRNA (nucleoside-2'-O-)-methyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
The isoprenylcysteine o-methyltransferase carries out carboyxl methylation of cleaved eukaryotic proteins that terminate in a CaaX motif. In Saccharomyces cerevisiae this methylation is carried out by Ste14p, an integral endoplasmic reticulum membrane protein. Ste14p is the founding member of the isoprenylcysteine carboxyl methyltransferase (ICMT) family, whose members share significant sequence homology.

Aminocyclopropane-1-carboxylic acid synthase is an enzyme that catalyzes the synthesis of 1-Aminocyclopropane-1-carboxylic acid (ACC), a precursor for ethylene, from S-Adenosyl methionine, an intermediate in the Yang cycle and activated methyl cycle and a useful molecule for methyl transfer. ACC synthase, like other PLP dependent enzymes, catalyzes the reaction through a quinonoid zwitterion intermediate and uses cofactor pyridoxal phosphate for stabilization.
In enzymology, an adenosylhomocysteine nucleosidase (EC 3.2.2.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a S-adenosylhomocysteine deaminase (EC 3.5.4.28) is an enzyme that catalyzes the chemical reaction

Adenosine kinase is an enzyme that catalyzes the transfer of gamma-phosphate from Adenosine triphosphate (ATP) to adenosine (Ado) leading to formation of Adenosine monophosphate (AMP). In addition to its well-studied role in controlling the cellular concentration of Ado, AdK also plays an important role in the maintenance of methylation reactions. All S-adenosylmethionine-dependent transmethylation reactions in cells lead to production of S-adenosylhomocysteine (SAH), which is cleaved by SAH hydrolase into Ado and homocysteine. The failure to efficiently remove these end products can result in buildup of SAH, which is a potent inhibitor of all transmethylation reactions. The disruption of AdK gene (-/-) in mice causes neonatal hepatic steatosis, a fatal condition characterized by rapid microvesicular fat infiltration, leading to early postnatal death. The liver was the main organ affected in these animals and in it the levels of adenine nucleotides were decreased, while those of SAH were elevated. Recently, missense mutations in the AdK gene in humans which result in AdK deficiency have also been shown to cause hypermethioninemia, encephalopathy and abnormal liver function.

Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.
Methyl halide transferase is an enzyme with systematic name S-adenosylmethionine:iodide methyltransferase. This enzyme catalyses the following chemical reaction