Chelation is a type of bonding of ions and molecules to metal ions. It involves the formation or presence of two or more separate coordinate bonds between a polydentate ligand and a single central metal atom. These ligands are called chelants, chelators, chelating agents, or sequestering agents. They are usually organic compounds, but this is not a necessity.

Medicinal or pharmaceutical chemistry is a scientific discipline at the intersection of chemistry and pharmacy involved with designing and developing pharmaceutical drugs. Medicinal chemistry involves the identification, synthesis and development of new chemical entities suitable for therapeutic use. It also includes the study of existing drugs, their biological properties, and their quantitative structure-activity relationships (QSAR).
A polycatenane is a chemical substance that, like polymers, is chemically constituted by a large number of units. These units are made up of concatenated rings into a chain-like structure.

In chemistry, a molecular knot is a mechanically interlocked molecular architecture that is analogous to a macroscopic knot. Naturally-forming molecular knots are found in organic molecules like DNA, RNA, and proteins. It is not certain that naturally occurring knots are evolutionarily advantageous to nucleic acids or proteins, though knotting is thought to play a role in the structure, stability, and function of knotted biological molecules. The mechanism by which knots naturally form in molecules, and the mechanism by which a molecule is stabilized or improved by knotting, is ambiguous. The study of molecular knots involves the formation and applications of both naturally occurring and chemically synthesized molecular knots. Applying chemical topology and knot theory to molecular knots allows biologists to better understand the structures and synthesis of knotted organic molecules.

Chemical biology is a scientific discipline between the fields of chemistry and biology. The discipline involves the application of chemical techniques, analysis, and often small molecules produced through synthetic chemistry, to the study and manipulation of biological systems. Although often confused with biochemistry, which studies the chemistry of biomolecules and regulation of biochemical pathways within and between cells, chemical biology remains distinct by focusing on the application of chemical tools to address biological questions.

In chemistry, an ionophore is a chemical species that reversibly binds ions. Many ionophores are lipid-soluble entities that transport ions across the cell membrane. Ionophores catalyze ion transport across hydrophobic membranes, such as liquid polymeric membranes or lipid bilayers found in the living cells or synthetic vesicles (liposomes). Structurally, an ionophore contains a hydrophilic center and a hydrophobic portion that interacts with the membrane.

Aptamers are short sequences of artificial DNA, RNA, XNA, or peptide that bind a specific target molecule, or family of target molecules. They exhibit a range of affinities, with variable levels of off-target binding and are sometimes classified as chemical antibodies. Aptamers and antibodies can be used in many of the same applications, but the nucleic acid-based structure of aptamers, which are mostly oligonucleotides, is very different from the amino acid-based structure of antibodies, which are proteins. This difference can make aptamers a better choice than antibodies for some purposes.

Endohedral fullerenes, also called endofullerenes, are fullerenes that have additional atoms, ions, or clusters enclosed within their inner spheres. The first lanthanum C60 complex called La@C60 was synthesized in 1985. The @ (at sign) in the name reflects the notion of a small molecule trapped inside a shell. Two types of endohedral complexes exist: endohedral metallofullerenes and non-metal doped fullerenes.

Pentetic acid or diethylenetriaminepentaacetic acid (DTPA) is an aminopolycarboxylic acid consisting of a diethylenetriamine backbone with five carboxymethyl groups. The molecule can be viewed as an expanded version of EDTA and is used similarly. It is a white solid with limited solubility in water.

TAP-associated glycoprotein, also known as tapasin or TAPBP, is a protein that in humans is encoded by the TAPBP gene.

Xiaoliang Sunney Xie is a Chinese biophysicist well known for his contributions to the fields of single-molecule biophysical chemistry, coherent Raman Imaging and single-molecule genomics. In 2023, Xie renounced his U.S. citizenship in order to reclaim his Chinese citizenship.

DOTA-TATE is an eight amino acid long peptide, with a covalently bonded DOTA bifunctional chelator.

Ammonia transporters are structurally related membrane transport proteins called Amt proteins in bacteria and plants, methylammonium/ammonium permeases (MEPs) in yeast, or Rhesus (Rh) proteins in chordates. In humans, the RhAG, RhBG, and RhCG Rhesus proteins constitute solute carrier family 42 whilst RhD and RhCE form the Rh blood group system. The three-dimensional structure of the ammonia transport protein AmtB from Escherichia coli has been determined by x-ray crystallography revealing a hydrophobic ammonia channel. The human RhCG ammonia transporter was found to have a similar ammonia-conducting channel structure. It was proposed that the erythrocyte Rh complex is a heterotrimer of RhAG, RhD, and RhCE subunits in which RhD and RhCE might play roles in anchoring the ammonia-conducting RhAG subunit to the cytoskeleton. Based on reconstitution experiments, purified RhCG subunits alone can function to transport ammonia. RhCG is required for normal acid excretion by the mouse kidney and epididymis.

The 3C-like protease (3CLpro) or main protease (Mpro), formally known as C30 endopeptidase or 3-chymotrypsin-like protease, is the main protease found in coronaviruses. It cleaves the coronavirus polyprotein at eleven conserved sites. It is a cysteine protease and a member of the PA clan of proteases. It has a cysteine-histidine catalytic dyad at its active site and cleaves a Gln–(Ser/Ala/Gly) peptide bond.
Topological inhibitors are rigid three-dimensional molecules of inorganic, organic, and hybrid compounds that form multicentered supramolecular interactions in vacant cavities of protein macromolecules and their complexes . Extensive surface and very diverse geometry make cage compounds with an encapsulated metal ion (clathrochelates) suitable for targeting both the active and allosteric sites of enzymes as well as the interfaces of their macromolecular complexes. An efficient structure- and concentration-dependent transcription inhibition in a model in vitro systems based on RNA and DNA polymerases by the iron(II) mono- and bis-clathrochelates at their submicro- and nanomolar concentrations, respectively, is observed in. Molecular docking and preincubation experiments suggested that these cage compounds form supramolecular assemblies with protein residues as well as with DNA and RNA. Thus, they are prospective precursors for the design of antiviral and anticancer drug candidates.
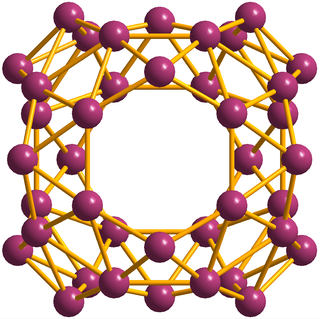
Borospherene (B40) is an electron-deficient cluster molecule containing 40 boron atoms. It bears similarities to other homoatomic cluster strucrures such as buckminsterfullerene (C60), stannaspherene, and plumbaspherene, but with a different symmetry. The first experimental evidence for borospherene was reported in July 2014, and is described in the journal Nature Chemistry. The molecule includes unusual hexagonal and heptagonal faces. Despite many calculation-based investigations into its structure and properties, a viable route for the synthesis and isolation of borospherene has yet to be established, and as a consequence it is still relatively poorly understood.

Macromolecular cages have three dimensional chambers surrounded by a molecular framework. Macromolecular cage architectures come in various sizes ranging from 1-50 nm and have varying topologies as well as functions. They can be synthesized through covalent bonding or self-assembly through non-covalent interactions. Most macromolecular cages that are formed through self-assembly are sensitive to pH, temperature, and solvent polarity.

The membrane (M) protein is an integral membrane protein that is the most abundant of the four major structural proteins found in coronaviruses. The M protein organizes the assembly of coronavirus virions through protein-protein interactions with other M protein molecules as well as with the other three structural proteins, the envelope (E), spike (S), and nucleocapsid (N) proteins.

The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid. The N protein is the most highly expressed of the four major coronavirus structural proteins. In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly. N also has additional functions in manipulating the cell cycle of the host cell. The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and COVID-19.

Spike (S) glycoprotein is the largest of the four major structural proteins found in coronaviruses. The spike protein assembles into trimers that form large structures, called spikes or peplomers, that project from the surface of the virion. The distinctive appearance of these spikes when visualized using negative stain transmission electron microscopy, "recalling the solar corona", gives the virus family its main name.

















