Related Research Articles

Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.
Deoxyribonuclease refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families, which differ in their substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include purifying proteins when extracted from prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Assays of DNase are emerging in the research field as well.

In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Mung bean nuclease is a nuclease derived from sprouts of the mung bean that removes nucleotides in a step-wise manner from single-stranded DNA molecules (ssDNA) and is used in biotechnological applications to remove such ssDNA from a mixture also containing double-stranded DNA (dsDNA). This enzyme is useful for transcript mapping, removal of single-stranded regions in DNA hybrids or single-stranded overhangs produced by restriction enzymes, etc. It has an activity similar to Nuclease S1, but it has higher specificity for single-stranded molecules.

NlaIII is a type II restriction enzyme isolated from Neisseria lactamica. As part of the restriction modification system, NlaIII is able to prevent foreign DNA from integrating into the host genome by cutting double stranded DNA into fragments at specific sequences. This results in further degradation of the fragmented foreign DNA and prevents it from infecting the host genome.
Type I site-specific deoxyribonuclease is an enzyme. This enzyme catalyses the following chemical reaction
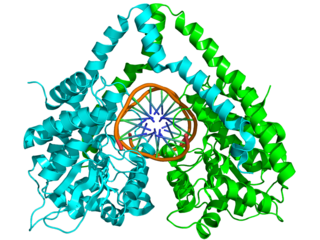
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.

EcoRV is a type II restriction endonuclease isolated from certain strains of Escherichia coli. It has the alternative name Eco32I.
Type II site-specific deoxyribonuclease is an enzyme. This enzyme catalyses the following chemical reaction
Type III site-specific deoxyribonuclease is an enzyme. This enzyme catalyses the following chemical reaction
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

Cassette mutagenesis is a type of site-directed mutagenesis that uses a short, double-stranded oligonucleotide sequence to replace a fragment of target DNA. It uses complementary restriction enzyme digest ends on the target DNA and gene cassette to achieve specificity. It is different from methods that use single oligonucleotide in that a single gene cassette can contain multiple mutations. Unlike many site directed mutagenesis methods, cassette mutagenesis also does not involve primer extension by DNA polymerase.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology, cell biology, and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.
References
- ↑ Theriault G, Roy PH, Howard KA, Benner JS, Brooks JE, Waters AF, Gingeras TR (December 1985). "Nucleotide sequence of the PaeR7 restriction/modification system and partial characterization of its protein products". Nucleic Acids Res. 13 (23): 8441–61. doi:10.1093/nar/13.23.8441. PMC 322144 . PMID 3001639.