A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
The restriction modification system is found in bacteria and archaea, and provides a defense against foreign DNA, such as that borne by bacteriophages.

In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zinc-finger nucleases to target unique sequences within complex genomes. By taking advantage of endogenous DNA repair machinery, these reagents can be used to precisely alter the genomes of higher organisms. Alongside CRISPR/Cas9 and TALEN, ZFN is a prominent tool in the field of genome editing.

The restriction endonuclease Fok1, naturally found in Flavobacterium okeanokoites, is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non sequence-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves the DNA at two locations, regardless of the nucleotide sequence at the cut site. The DNA is cut 9 nucleotides downstream of the motif on the forward strand, and 13 nucleotides downstream of the motif on the reverse strand, producing two sticky ends with 4-bp overhangs.
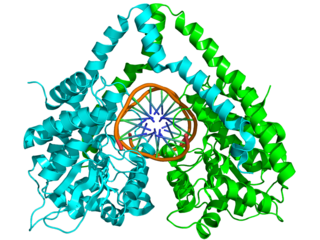
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.

HaeIII is one of many restriction enzymes (endonucleases) a type of prokaryotic DNA that protects organisms from unknown, foreign DNA. It is a restriction enzyme used in molecular biology laboratories. It was the third endonuclease to be isolated from the Haemophilus aegyptius bacteria. The enzyme's recognition site—the place where it cuts DNA molecules—is the GGCC nucleotide sequence which means it cleaves DNA at the site 5′-GG/CC-3. The recognition site is usually around 4-8 bps.This enzyme's gene has been sequenced and cloned. This is done to make DNA fragments in blunt ends. HaeIII is not effective for single stranded DNA cleavage.
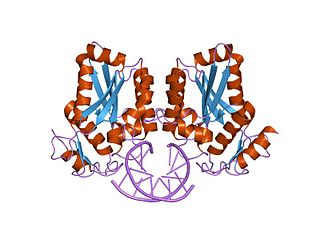
BamHI is a type II restriction endonuclease, having the capacity for recognizing short sequences of DNA and specifically cleaving them at a target site. This exhibit focuses on the structure-function relations of BamHI as described by Newman, et al. (1995). BamHI binds at the recognition sequence 5'-GGATCC-3', and cleaves these sequences just after the 5'-guanine on each strand. This cleavage results in sticky ends which are 4 bp long. In its unbound form, BamHI displays a central b sheet, which resides in between α-helices.

The homing endonucleases are a collection of endonucleases encoded either as freestanding genes within introns, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele frequency at greater than Mendelian rates.

Nuclease S1 is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

BglII is a type II restriction endonuclease isolated from certain strains of Bacillus globigii.
Chimeric nucleases are an example of engineered proteins which must comprise a DNA-binding domain to give sequence specificity and a nuclease domain for DNA cleavage.
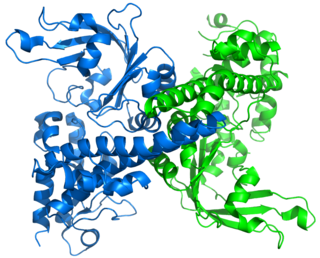
Restriction endonuclease (REase) EcoRII is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.
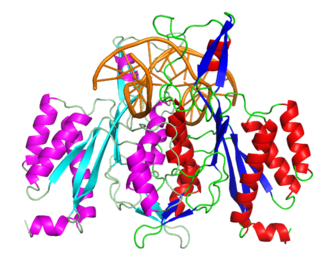
EcoRI is a restriction endonuclease enzyme isolated from species E. coli. It is a restriction enzyme that cleaves DNA double helices into fragments at specific sites, and is also a part of the restriction modification system. The Eco part of the enzyme's name originates from the species from which it was isolated - "E" denotes generic name which is "Escherichia" and "co" denotes species name, "coli" - while the R represents the particular strain, in this case RY13, and the I denotes that it was the first enzyme isolated from this strain.
Since antiretroviral therapy requires a lifelong treatment regimen, research to find more permanent cures for HIV infection is currently underway. It is possible to synthesize zinc finger nucleotides with zinc finger components that selectively bind to specific portions of DNA. Conceptually, targeting and editing could focus on host cellular co-receptors for HIV or on proviral HIV DNA.











