A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
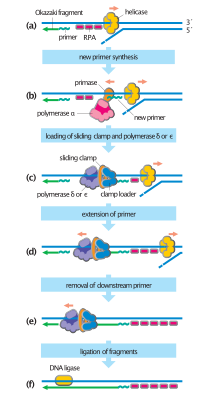
DnaG is a bacterial DNA primase and is encoded by the dnaG gene. The enzyme DnaG, and any other DNA primase, synthesizes short strands of RNA known as oligonucleotides during DNA replication. These oligonucleotides are known as primers because they act as a starting point for DNA synthesis. DnaG catalyzes the synthesis of oligonucleotides that are 10 to 60 nucleotides long, however most of the oligonucleotides synthesized are 11 nucleotides. These RNA oligonucleotides serve as primers, or starting points, for DNA synthesis by bacterial DNA polymerase III. DnaG is important in bacterial DNA replication because DNA polymerase cannot initiate the synthesis of a DNA strand, but can only add nucleotides to a preexisting strand. DnaG synthesizes a single RNA primer at the origin of replication. This primer serves to prime leading strand DNA synthesis. For the other parental strand, the lagging strand, DnaG synthesizes an RNA primer every few kilobases (kb). These primers serve as substrates for the synthesis of Okazaki fragments.

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.

Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.

Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.

Replication protein A 70 kDa DNA-binding subunit is a protein that in humans is encoded by the RPA1 gene.

DNA topoisomerase 2-binding protein 1 (TOPBP1) is a scaffold protein that in humans is encoded by the TOPBP1 gene.

RAD52 homolog , also known as RAD52, is a protein which in humans is encoded by the RAD52 gene.

Meiotic recombination protein DMC1/LIM15 homolog is a protein that in humans is encoded by the DMC1 gene.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.
Shelterin is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop).
Single-strand DNA-binding protein (SSB) is a protein found in Escherichia coli bacteria, that binds to single-stranded regions of deoxyribonucleic acid (DNA). Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination, and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes.
DNA polymerase epsilon is a member of the DNA polymerase family of enzymes found in eukaryotes. It is composed of the following four subunits: POLE, POLE2, POLE3, and POLE4. Recent evidence suggests that it plays a major role in leading strand DNA synthesis and nucleotide and base excision repair.

Replication protein A 32 kDa subunit is a protein that in humans is encoded by the RPA2 gene.
Replication protein A 14 kDa subunit is a protein that in humans is encoded by the RPA3 gene.

DNA end resection, also called 5′–3′ degradation, is a biochemical process where the blunt end of a section of double-stranded DNA (dsDNA) is modified by cutting away some nucleotides from the 5' end to produce a 3' single-stranded sequence. The presence of a section of single-stranded DNA (ssDNA) allows the broken end of the DNA to line up accurately with a matching sequence, so that it can be accurately repaired.

In molecular biology, the OB-fold is a small protein structural motif observed in different proteins that bind oligonucleotides or oligosaccharides. It was originally identified in 1993 in four unrelated proteins: staphylococcal nuclease, anticodon binding domain of aspartyl-tRNA synthetase, and the B-subunits of heat-labile enterotoxin and verotoxin-1. Since then it has been found in multiple proteins many of which are involved in genome stability. This fold is often described as a Greek key motif.