
In immunology, a Fc receptor is a protein found on the surface of certain cells – including, among others, B lymphocytes, follicular dendritic cells, natural killer cells, macrophages, neutrophils, eosinophils, basophils, human platelets, and mast cells – that contribute to the protective functions of the immune system. Its name is derived from its binding specificity for a part of an antibody known as the Fc region. Fc receptors bind to antibodies that are attached to infected cells or invading pathogens. Their activity stimulates phagocytic or cytotoxic cells to destroy microbes, or infected cells by antibody-mediated phagocytosis or antibody-dependent cell-mediated cytotoxicity. Some viruses such as flaviviruses use Fc receptors to help them infect cells, by a mechanism known as antibody-dependent enhancement of infection.
Co-stimulation is a secondary signal which immune cells rely on to activate an immune response in the presence of an antigen-presenting cell. In the case of T cells, two stimuli are required to fully activate their immune response. During the activation of lymphocytes, co-stimulation is often crucial to the development of an effective immune response. Co-stimulation is required in addition to the antigen-specific signal from their antigen receptors.
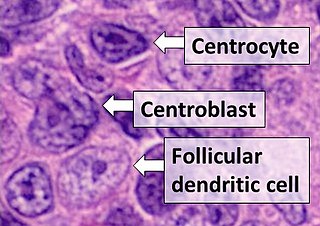
Follicular dendritic cells (FDC) are cells of the immune system found in primary and secondary lymph follicles of the B cell areas of the lymphoid tissue. Unlike dendritic cells (DC), FDCs are not derived from the bone-marrow hematopoietic stem cell, but are of mesenchymal origin. Possible functions of FDC include: organizing lymphoid tissue's cells and microarchitecture, capturing antigen to support B cell, promoting debris removal from germinal centers, and protecting against autoimmunity. Disease processes that FDC may contribute include primary FDC-tumor, chronic inflammatory conditions, HIV-1 infection development, and neuroinvasive scrapie.

The Cluster of differentiation 80 is a B7, type I membrane protein in the immunoglobulin superfamily, with an extracellular immunoglobulin constant-like domain and a variable-like domain required for receptor binding. It is closely related to CD86, another B7 protein (B7-2), and often works in tandem. Both CD80 and CD86 interact with costimulatory receptors CD28, CTLA-4 (CD152) and the p75 neurotrophin receptor.
An immunoreceptor tyrosine-based inhibitory motif (ITIM), is a conserved sequence of amino acids that is found intracellularly in the cytoplasmic domains of many inhibitory receptors of the non-catalytic tyrosine-phosphorylated receptor family found on immune cells. These immune cells include T cells, B cells, NK cells, dendritic cells, macrophages and mast cells. ITIMs have similar structures of S/I/V/LxYxxI/V/L, where x is any amino acid, Y is a tyrosine residue that can be phosphorylated, S is the amino acide Serine, I is the amino acid Isoleucine, and V is the amino acid Valine. ITIMs recruit SH2 domain-containing phosphatases, which inhibit cellular activation. ITIM-containing receptors often serve to target Immunoreceptor tyrosine-based activation motif(ITAM)-containing receptors, resulting in an innate inhibition mechanism within cells. ITIM bearing receptors have important role in regulation of immune system allowing negative regulation at different levels of the immune response.

CD5 is a cluster of differentiation expressed on the surface of T cells and in a subset of murine B cells known as B-1a. The expression of this receptor in human B cells has been a controversial topic and to date there is no consensus regarding the role of this receptor as a marker of human B cells. B-1 cells have limited diversity of their B-cell receptor due to their lack of the enzyme terminal deoxynucleotidyl transferase (TdT) and are potentially self-reactive. CD5 serves to mitigate activating signals from the BCR so that the B-1 cells can only be activated by very strong stimuli and not by normal tissue proteins. CD5 was used as a T-cell marker until monoclonal antibodies against CD3 were developed.

Programmed death-ligand 1 (PD-L1) also known as cluster of differentiation 274 (CD274) or B7 homolog 1 (B7-H1) is a protein that in humans is encoded by the CD274 gene.

Leukocyte immunoglobulin-like receptor subfamily B member 1 is a protein that in humans is encoded by the LILRB1 gene.

CD83 is a human protein encoded by the CD83 gene.

CD69 is a human transmembrane C-Type lectin protein encoded by the CD69 gene. It is an early activation marker that is expressed in hematopoietic stem cells, T cells, and many other cell types in the immune system. It is also implicated in T cell differentiation as well as lymphocyte retention in lymphoid organs.

CD244 also known as 2B4 or SLAMF4 is a protein that in humans is encoded by the CD244 gene.

CD226, PTA1 or DNAM-1 is a ~65 kDa immunoglobulin-like transmembrane glycoprotein expressed on the surface of natural killer cells, NK T cell, B cells, dendritic cells, hematopoietic precursor cells, platelets, monocytes and T cells.

OX-2 membrane glycoprotein, also named CD200 is a human protein encoded by the CD200 gene. CD200 gene is in human located on chromosome 3 in proximity to genes encoding other B7 proteins CD80/CD86. In mice CD200 gene is on chromosome 16.

Hepatitis A virus cellular receptor 2 (HAVCR2), also known as T-cell immunoglobulin and mucin-domain containing-3 (TIM-3), is a protein that in humans is encoded by the HAVCR2 (TIM-3)gene. HAVCR2 was first described in 2002 as a cell surface molecule expressed on IFNγ producing CD4+ Th1 and CD8+ Tc1 cells. Later, the expression was detected in Th17 cells, regulatory T-cells, and innate immune cells. HAVCR2 receptor is a regulator of the immune response.

Killer cell lectin-like receptor subfamily G member 1 is a protein that in humans is encoded by the KLRG1 gene.
Fc fragment of IgG receptor IIb is a low affinity inhibitory receptor for the Fc region of immunoglobulin gamma (IgG). FCGR2B participates in the phagocytosis of immune complexes and in the regulation of antibody production by B lymphocytes.

Fc fragment of IgA receptor (FCAR) is a human gene that codes for the transmembrane receptor FcαRI, also known as CD89. FcαRI binds the heavy-chain constant region of Immunoglobulin A (IgA) antibodies. FcαRI is present on the cell surface of myeloid lineage cells, including neutrophils, monocytes, macrophages, and eosinophils, though it is notably absent from intestinal macrophages and does not appear on mast cells. FcαRI plays a role in both pro- and anti-inflammatory responses depending on the state of IgA bound. Inside-out signaling primes FcαRI in order for it to bind its ligand, while outside-in signaling caused by ligand binding depends on FcαRI association with the Fc receptor gamma chain.

NKG2D is an activating receptor (transmembrane protein) belonging to the NKG2 family of C-type lectin-like receptors. NKG2D is encoded by KLRK1 (killer cell lectin like receptor K1) gene which is located in the NK-gene complex (NKC) situated on chromosome 6 in mice and chromosome 12 in humans. In mice, it is expressed by NK cells, NK1.1+ T cells, γδ T cells, activated CD8+ αβ T cells and activated macrophages. In humans, it is expressed by NK cells, γδ T cells and CD8+ αβ T cells. NKG2D recognizes induced-self proteins from MIC and RAET1/ULBP families which appear on the surface of stressed, malignant transformed, and infected cells.

Immune checkpoints are regulators of the immune system. These pathways are crucial for self-tolerance, which prevents the immune system from attacking cells indiscriminately. However, some cancers can protect themselves from attack by stimulating immune checkpoint targets.

CD28 family receptors are a group of regulatory cell surface receptors expressed on immune cells. The CD28 family in turn is a subgroup of the immunoglobulin superfamily.























