
Structural biology is a field that is many centuries old which, and as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization. Early structural biologists throughout the 19th and early 20th centuries were primarily only able to study structures to the limit of the naked eye's visual acuity and through primitive magnifying glasses and light microscopes. In the last half of the 20th century through the discovery and development of the electron microscope the field was revolutionized, as now scientists could image the structure of cells, organelles, and large extracellular matrix proteins over multiple length scales and starting at several nanometers resolution by the end of the century. In the 21st century, the field saw another drastic revolution with the development of more coherent electron sources, aberration correction for electron microscopes, and reconstruction software that enabled the success of high resolution cryo-electron tomography which permits the study of individual proteins and molecular complexes in 3-dimensions at angstroms resolution. Additional tools were also developed and refined around this time to study the properties of the extracellular matrix including atomic-force microscopy, focused ion beam (FIB) and FIB-SEM slice and view, X-ray computed tomography, and more recently deep-learning tools for segmentation of structures in 3-dimensions. With the development of the prior cryo-electron tomography the field of structural biology became much larger and also became a branch of molecular biology, biochemistry, and biophysics concerned with the molecular structure of biological macromolecules, how they acquire the structures they have, and how alterations in their structures affect their function. This subject is of great interest to biologists because macromolecules carry out most of the functions of cells, and it is only by coiling into specific three-dimensional shapes that they are able to perform these functions. This architecture, the "tertiary structure" of molecules, depends in a complicated way on each molecule's basic composition, or "primary structure." At lower resolutions, tools such as FIB-SEM tomography have allowed for greater understanding of cells and their organelles in 3-dimensions, and how each hierarchical level of various extracellular matrices contributes to function. In the past few years it has become possible for highly accurate physical molecular models to complement the in silico study of biological structures. Examples of these models can be found in the Protein Data Bank. Computational techniques like Molecular Dynamics simulations can be used in conjunction with empirical structure determination strategies to extend and study protein structure, conformation and function.
Bacteriorhodopsin is a protein used by Archaea, most notably by haloarchaea, a class of the Euryarchaeota. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell. The resulting proton gradient is subsequently converted into chemical energy.
Electron crystallography is a method to determine the arrangement of atoms in solids using a transmission electron microscope (TEM).

Transmission electron cryomicroscopy (CryoTEM), commonly known as cryo-EM, is a form of cryogenic electron microscopy, more specifically a type of transmission electron microscopy (TEM) where the sample is studied at cryogenic temperatures. Cryo-EM is gaining popularity in structural biology.

Electron cryotomography (CryoET) is an imaging technique used to produce high-resolution (~1–4 nm) three-dimensional views of samples, often biological macromolecules and cells. CryoET is a specialized application of transmission electron cryomicroscopy (CryoTEM) in which samples are imaged as they are tilted, resulting in a series of 2D images that can be combined to produce a 3D reconstruction, similar to a CT scan of the human body. In contrast to other electron tomography techniques, samples are imaged under cryogenic conditions. For cellular material, the structure is immobilized in non-crystalline ("vitreous") ice and allowing them to be imaged without dehydration or chemical fixation, which could otherwise disrupt or distort biological structures.

Richard Henderson is a Scottish molecular biologist and biophysicist and pioneer in the field of electron microscopy of biological molecules. Henderson shared the Nobel Prize in Chemistry in 2017 with Jacques Dubochet and Joachim Frank.

Eva Nogales is a biophysicist at the Lawrence Berkeley National Laboratory and a Professor at the University of California, Berkeley, where she served head of the Division of Biochemistry, Biophysics and Structural Biology of the Department of Molecular and Cell Biology (2015-2020). She is a Howard Hughes Medical Institute investigator.
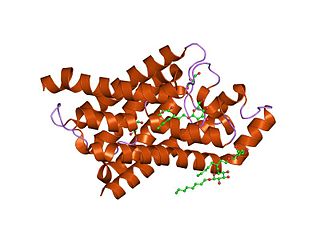
Major intrinsic proteins comprise a large superfamily of transmembrane protein channels that are grouped together on the basis of homology. The MIP superfamily includes three subfamilies: aquaporins, aquaglyceroporins and S-aquaporins.
- The aquaporins (AQPs) are water selective.
- The aquaglyceroporins are permeable to water, but also to other small uncharged molecules such as glycerol.
- The third subfamily, with little conserved amino acid sequences around the NPA boxes, include 'superaquaporins' (S-aquaporins).
Resolution in terms of electron density is a measure of the resolvability in the electron density map of a molecule. In X-ray crystallography, resolution is the highest resolvable peak in the diffraction pattern, while resolution in cryo-electron microscopy is a frequency space comparison of two halves of the data, which strives to correlate with the X-ray definition.

Protein crystallization is the process of formation of a regular array of individual protein molecules stabilized by crystal contacts. If the crystal is sufficiently ordered, it will diffract. Some proteins naturally form crystalline arrays, like aquaporin in the lens of the eye.
Chikashi Toyoshima (豊島 近, Toyoshima Chikashi, born July 17,1954) is a Japanese biophysicist. His research interest only focus on two proteins: the Ca2+-ATPase of muscle sarcoplasmic reticulum, and the Na+, K+-ATPase expressed in all animal cells. He is a professor of University of Tokyo and the Foreign Associate of the National Academy of Sciences, USA. His research about the Ca2+-ATPase started in 1989. In the next few years, he and his colleagues obtained a series of images of Ca2+-ATPase at the revolution of Atomic-level in the world for the first time. By the x-ray crystallography, cryo-EM and other methods, he has determined the crystal structures of ten intermediates of Ca2+-ATPase. On September 10, 2015, The Royal Swedish Academy of Sciences awarded him and Poul Nissen the Gregori Aminoff Prize of 2016 for their fundamental contributions to understanding the structural basis for ATP-driven translocation of ions across membrane.
In crystallography, direct methods is a set of techniques used for structure determination using diffraction data and a priori information. It is a solution to the crystallographic phase problem, where phase information is lost during a diffraction measurement. Direct methods provides a method of estimating the phase information by establishing statistical relationships between the recorded amplitude information and phases of strong reflections.

Cryogenic electron microscopy (cryo-EM) is a cryomicroscopy technique applied on samples cooled to cryogenic temperatures. For biological specimens, the structure is preserved by embedding in an environment of vitreous ice. An aqueous sample solution is applied to a grid-mesh and plunge-frozen in liquid ethane or a mixture of liquid ethane and propane. While development of the technique began in the 1970s, recent advances in detector technology and software algorithms have allowed for the determination of biomolecular structures at near-atomic resolution. This has attracted wide attention to the approach as an alternative to X-ray crystallography or NMR spectroscopy for macromolecular structure determination without the need for crystallization.

Sjors Hendrik Willem ScheresFRS is a Dutch scientist at the MRC Laboratory of Molecular Biology Cambridge, UK.
Microcrystal electron diffraction, or MicroED, is a CryoEM method that was developed by the Gonen laboratory in late 2013 at the Janelia Research Campus of the Howard Hughes Medical Institute. MicroED is a form of electron crystallography where thin 3D crystals are used for structure determination by electron diffraction.

Kiyoshi Nagai was a Japanese structural biologist at the MRC Laboratory of Molecular Biology Cambridge, UK. He was known for his work on the mechanism of RNA splicing and structures of the spliceosome.

Serial femtosecond crystallography (SFX) is a form of X-ray crystallography developed for use at X-ray free-electron lasers (XFELs). Single pulses at free-electron lasers are bright enough to generate resolvable Bragg diffraction from sub-micron crystals. However, these pulses also destroy the crystals, meaning that a full data set involves collecting diffraction from many crystals. This method of data collection is referred to as serial, referencing a row of crystals streaming across the X-ray beam, one at a time.
Leonid A. Sazanov is a professor at the Institute of Science and Technology Austria (IST). Sazanov research explores the structure and function of large membrane protein complexes from the domain of bioenergetics. These molecular machines interconvert redox energy and proton motive force across biological membranes using a variety of mechanisms.
Hosea Nelson is an American chemist who is a professor at California Institute of Technology. His research investigates the design and total synthesis of complex molecules. He was a finalist for the 2021 Blavatnik Awards for Young Scientists.











