
Structural biology, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization.
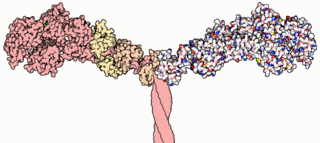
Myosins are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.

Inositol trisphosphate receptor (InsP3R) is a membrane glycoprotein complex acting as a Ca2+ channel activated by inositol trisphosphate (InsP3). InsP3R is very diverse among organisms, and is necessary for the control of cellular and physiological processes including cell division, cell proliferation, apoptosis, fertilization, development, behavior, learning and memory. Inositol triphosphate receptor represents a dominant second messenger leading to the release of Ca2+ from intracellular store sites. There is strong evidence suggesting that the InsP3R plays an important role in the conversion of external stimuli to intracellular Ca2+ signals characterized by complex patterns relative to both space and time, such as Ca2+ waves and oscillations.

The tau proteins form a group of six highly soluble protein isoforms produced by alternative splicing from the gene MAPT. They have roles primarily in maintaining the stability of microtubules in axons and are abundant in the neurons of the central nervous system (CNS), where the cerebral cortex has the highest abundance. They are less common elsewhere but are also expressed at very low levels in CNS astrocytes and oligodendrocytes.

Transmission electron cryomicroscopy (CryoTEM), commonly known as cryo-EM, is a form of cryogenic electron microscopy, more specifically a type of transmission electron microscopy (TEM) where the sample is studied at cryogenic temperatures. Cryo-EM, specifically 3-dimensional electron microscopy (3DEM), is gaining popularity in structural biology.

Hugh Esmor Huxley MBE FRS was a British molecular biologist who made important discoveries in the physiology of muscle. He was a graduate in physics from Christ's College, Cambridge. However, his education was interrupted for five years by the Second World War, during which he served in the Royal Air Force. His contribution to development of radar earned him an MBE.

Venkatraman Ramakrishnan is a British-American structural biologist. He shared the 2009 Nobel Prize in Chemistry with Thomas A. Steitz and Ada Yonath for research on the structure and function of ribosomes.

The Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) is a research institute in Cambridge, England, involved in the revolution in molecular biology which occurred in the 1950–60s. Since then it has remained a major medical research laboratory at the forefront of scientific discovery, dedicated to improving the understanding of key biological processes at atomic, molecular and cellular levels using multidisciplinary methods, with a focus on using this knowledge to address key issues in human health.

Richard Henderson is a British molecular biologist and biophysicist and pioneer in the field of electron microscopy of biological molecules. Henderson shared the Nobel Prize in Chemistry in 2017 with Jacques Dubochet and Joachim Frank. "Thanks to his work, we can look at individual atoms of living nature, thanks to cryo-electron microscopes we can see details without destroying samples, and for this he won the Nobel Prize in Chemistry."
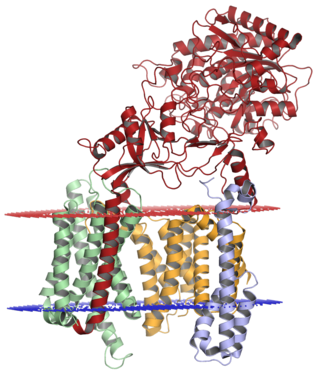
Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44.
Resolution in the context of structural biology is the ability to distinguish the presence or absence of atoms or groups of atoms in a biomolecular structure. Usually, the structure originates from methods such as X-ray crystallography, electron crystallography, or cryo-electron microscopy. The resolution is measured of the "map" of the structure produced from experiment, where an atomic model would then be fit into. Due to their different natures and interactions with matter, in X-ray methods the map produced is of the electron density of the system, whereas in electron methods the map is of the electrostatic potential of the system. In both cases, atomic positions are assumed similarly.
Chikashi Toyoshima (豊島 近, Toyoshima Chikashi, born July 17, 1954) is a Japanese biophysicist. His research focuses on two proteins: the Ca2+-ATPase of muscle sarcoplasmic reticulum, and the Na+, K+-ATPase expressed in all animal cells. He is a professor at the University of Tokyo and the Foreign Associate of the National Academy of Sciences, USA. Toyoshima's research about the Ca2+-ATPase started in 1989. In the next few years, he and his colleagues obtained the world's first series of images of Ca2+-ATPase at the atomic level. Via x-ray crystallography, cryo-EM and other methods, he has determined the crystal structures of ten intermediates of Ca2+-ATPase. On September 10, 2015, The Royal Swedish Academy of Sciences awarded him and Poul Nissen the Gregori Aminoff Prize of 2016 for their fundamental contributions to understanding the structural basis for ATP-driven translocation of ions across membranes.

Cryogenic electron microscopy (cryo-EM) is a cryomicroscopy technique applied on samples cooled to cryogenic temperatures. For biological specimens, the structure is preserved by embedding in an environment of vitreous ice. An aqueous sample solution is applied to a grid-mesh and plunge-frozen in liquid ethane or a mixture of liquid ethane and propane. While development of the technique began in the 1970s, recent advances in detector technology and software algorithms have allowed for the determination of biomolecular structures at near-atomic resolution. This has attracted wide attention to the approach as an alternative to X-ray crystallography or NMR spectroscopy for macromolecular structure determination without the need for crystallization.

Tamir Gonen is an American structural biochemist and membrane biophysicist best known for his contributions to structural biology of membrane proteins, membrane biochemistry and electron cryo-microscopy (cryoEM) particularly in electron crystallography of 2D crystals and for the development of 3D electron crystallography from microscopic crystals known as MicroED. Gonen is an Investigator of the Howard Hughes Medical Institute, a professor at the University of California, Los Angeles, the founding director of the MicroED Imaging Center at UCLA and a Member of the Royal Society of New Zealand.

Andrew P. Carter is a British structural biologist who works at the Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) in Cambridge, UK. He is known for his work on the microtubule motor dynein.

Kiyoshi Nagai was a Japanese structural biologist at the MRC Laboratory of Molecular Biology Cambridge, UK. He was known for his work on the mechanism of RNA splicing and structures of the spliceosome.
Leonid A. Sazanov is a professor at the Institute of Science and Technology Austria (ISTA). Sazanov research explores the structure and function of large membrane protein complexes from the domain of bioenergetics. These molecular machines interconvert redox energy and proton motive force across biological membranes using a variety of mechanisms.

Jan Steyaert is a Belgian bioengineer and molecular biologist. He started his career as an enzymologist but the Steyaertlab is best known for pioneering work on (engineered) nanobodies for applications in structural biology, omics and drug design. He is full professor and teaches biochemistry at the Vrije Universiteit Brussel and Director of the VIB-VUB Center for Structural Biology, one of the Research Centers of the Vlaams Instituut voor Biotechnologie (VIB). He was involved in the foundation of three spin-off companies: Ablynx, Biotalys, and Confo Therapeutics.

Stefan Raunser is a German scientist and structural biologist specializing in membrane proteins, the cytoskeleton, toxins, and sarcomere structural biochemistry. Since 2014, he has been a director at the Max Planck Institute of Molecular Physiology in Dortmund, Germany.

Tanmay A. M. Bharat is a programme leader in the Structural Studies Division of the MRC Laboratory of Molecular Biology. He and his group use electron tomography, together with several structural and cell biology methods to study the cell surfaces of bacteria and archaea. His work has increased the understanding of how surface molecules help in the formation of multicellular communities of prokaryotes, examples of which include biofilms and microbiomes. He has been awarded several prizes and fellowships for his work.

















