
Epoxide hydrolases (EHs), also known as epoxide hydratases, are enzymes that metabolize compounds that contain an epoxide residue; they convert this residue to two hydroxyl residues through an epoxide hydrolysis reaction to form diol products. Several enzymes possess EH activity. Microsomal epoxide hydrolase, soluble epoxide hydrolase, and the more recently discovered but not as yet well defined functionally, epoxide hydrolase 3 (EH3) and epoxide hydrolase 4 (EH4) are structurally closely related isozymes. Other enzymes with epoxide hydrolase activity include leukotriene A4 hydrolase, Cholesterol-5,6-oxide hydrolase, MEST (gene) (Peg1/MEST), and Hepoxilin-epoxide hydrolase. The hydrolases are distinguished from each other by their substrate preferences and, directly related to this, their functions.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.
In enzymology, a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate reductase (EC 1.3.1.40) is an enzyme that catalyzes the chemical reaction
In enzymology, a pyrroloquinoline-quinone synthase (EC 1.3.3.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a terephthalate 1,2-cis-dihydrodiol dehydrogenase (EC 1.3.1.61) is an enzyme that catalyzes the chemical reaction:
In enzymology, a 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione 4,5-dioxygenase (EC 1.13.11.25) is an enzyme that catalyzes the chemical reaction
Biphenyl-2,3-diol 1,2-dioxygenase (EC 1.13.11.39) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2,6-dioxo-6-phenylhexa-3-enoate hydrolase (EC 3.7.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2-hydroxymuconate-semialdehyde hydrolase (EC 3.7.1.9) is an enzyme that catalyzes the chemical reaction
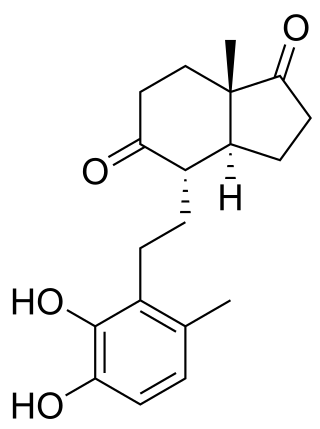
3,4-DHSA is an organic compound which is the intermediate product of the metabolism of cholesterol, by the bacteria most commonly responsible for tuberculosis. 3,4-DHSA is an acronym for 3,4-dihydroxy-9,10-seco-androst-1,3,5(10)-triene-9,17-dione, the official name of this substance. It is classified as a secosteroid, since one of the four rings of cholesterol from which it is derived is broken.
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Hydroquinone 1,2-dioxygenase (EC 1.13.11.66, hydroquinone dioxygenase) is an enzyme with systematic name benzene-1,4-diol:oxygen 1,2-oxidoreductase (decyclizing). This enzyme catalyses the following chemical reaction
Cholest-4-en-3-one 26-monooxygenase (EC 1.14.13.141, CYP125, CYP125A1, cholest-4-en-3-one 27-monooxygenase) is an enzyme with systematic name cholest-4-en-3-one,NADH:oxygen oxidoreductase (26-hydroxylating). This enzyme catalyses the following chemical reaction
3-ketosteroid 9alpha-monooxygenase (EC 1.14.13.142, KshAB, 3-ketosteroid 9alpha-hydroxylase) is an enzyme with systematic name androsta-1,4-diene-3,17-dione,NADH:oxygen oxidoreductase (9alpha-hydroxylating). This enzyme catalyses the following chemical reaction
3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase (EC 1.14.14.12, HsaA) is an enzyme with systematic name 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione,FMNH2:oxygen oxidoreductase. This enzyme catalyses the following chemical reaction:
2-hydroxy-6-oxo-6-(2-aminophenyl)hexa-2,4-dienoate hydrolase (EC 3.7.1.13, CarC) is an enzyme with systematic name (2E,4E)-6-(2-aminophenyl)-2-hydroxy-6-oxohexa-2,4-dienoate acylhydrolase. This enzyme catalyses the following chemical reaction
2-hydroxy-6-oxonona-2,4-dienedioate hydrolase (EC 3.7.1.14, mhpC (gene)) is an enzyme with systematic name (2Z,4E)-2-hydroxy-6-oxona-2,4-dienedioate succinylhydrolase. This enzyme catalyses the following chemical reaction:
- (2Z,4E)-2-hydroxy-6-oxonona-2,4-diene-1,9-dioate + H2O (2Z)-2-hydroxypenta-2,4-dienoate + succinate
- (2Z,4E,7E)-2-hydroxy-6-oxonona-2,4,7-triene-1,9-dioate + H2O (2Z)-2-hydroxypenta-2,4-dienoate + fumarate
2-hydroxyhexa-2,4-dienoate hydratase (EC 4.2.1.132, tesE (gene), hsaE (gene)) is an enzyme with systematic name 4-hydroxy-2-oxohexanoate hydro-lyase ((2Z,4Z)-2-hydroxyhexa-2,4-dienoate-forming). This enzyme catalyses the following chemical reaction



