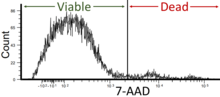
Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles.

In biochemistry, immunostaining is any use of an antibody-based method to detect a specific protein in a sample. The term "immunostaining" was originally used to refer to the immunohistochemical staining of tissue sections, as first described by Albert Coons in 1941. However, immunostaining now encompasses a broad range of techniques used in histology, cell biology, and molecular biology that use antibody-based staining methods.
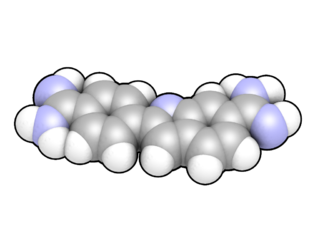
A fluorophore is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds.
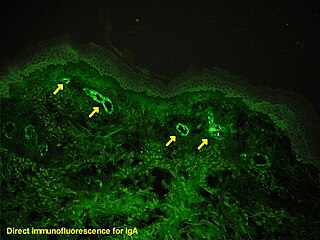
Immunofluorescence is a technique used for light microscopy with a fluorescence microscope and is used primarily on biological samples. This technique uses the specificity of antibodies to their antigen to target fluorescent dyes to specific biomolecule targets within a cell, and therefore allows visualization of the distribution of the target molecule through the sample. The specific region an antibody recognizes on an antigen is called an epitope. There have been efforts in epitope mapping since many antibodies can bind the same epitope and levels of binding between antibodies that recognize the same epitope can vary. Additionally, the binding of the fluorophore to the antibody itself cannot interfere with the immunological specificity of the antibody or the binding capacity of its antigen. Immunofluorescence is a widely used example of immunostaining and is a specific example of immunohistochemistry. This technique primarily makes use of fluorophores to visualise the location of the antibodies.
A total internal reflection fluorescence microscope (TIRFM) is a type of microscope with which a thin region of a specimen, usually less than 200 nanometers can be observed.

A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image.
DAPI, or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability.

Hoechst stains are part of a family of blue fluorescent dyes used to stain DNA. These bis-benzimides were originally developed by Hoechst AG, which numbered all their compounds so that the dye Hoechst 33342 is the 33,342nd compound made by the company. There are three related Hoechst stains: Hoechst 33258, Hoechst 33342, and Hoechst 34580. The dyes Hoechst 33258 and Hoechst 33342 are the ones most commonly used and they have similar excitation–emission spectra.

Two-photon excitation microscopy is a fluorescence imaging technique that is particularly well-suited to image scattering living tissue of up to about one millimeter in thickness. Unlike traditional fluorescence microscopy, where the excitation wavelength is shorter than the emission wavelength, two-photon excitation requires simultaneous excitation by two photons with longer wavelength than the emitted light. The laser is focused onto a specific location in the tissue and scanned across the sample to sequentially produce the image. Due to the non-linearity of two-photon excitation, mainly fluorophores in the micrometer-sized focus of the laser beam are excited, which results in the spatial resolution of the image. This contrasts with confocal microscopy, where the spatial resolution is produced by the interaction of excitation focus and the confined detection with a pinhole.

Propidium iodide is a fluorescent intercalating agent that can be used to stain cells and nucleic acids. PI binds to DNA by intercalating between the bases with little or no sequence preference. When in an aqueous solution, PI has a fluorescent excitation maximum of 493 nm (blue-green), and an emission maximum of 636 nm (red). After binding DNA, the quantum yield of PI is enhanced 20-30 fold, and the excitation/emission maximum of PI is shifted to 535 nm (green) / 617 nm (orange-red). Propidium iodide is used as a DNA stain in flow cytometry to evaluate cell viability or DNA content in cell cycle analysis, or in microscopy to visualize the nucleus and other DNA-containing organelles. Propidium Iodide is not membrane-permeable, making it useful to differentiate necrotic, apoptotic and healthy cells based on membrane integrity. PI also binds to RNA, necessitating treatment with nucleases to distinguish between RNA and DNA staining. PI is widely used in fluorescence staining and visualization of the plant cell wall.

Texas Red or sulforhodamine 101 acid chloride is a red fluorescent dye, used in histology for staining cell specimens, for sorting cells with fluorescent-activated cell sorting machines, in fluorescence microscopy applications, and in immunohistochemistry. Texas Red fluoresces at about 615 nm, and the peak of its absorption spectrum is at 589 nm. The powder is dark purple. Solutions can be excited by a dye laser tuned to 595-605 nm, or less efficiently a krypton laser at 567 nm. The absorption extinction coefficient at 596 nm is about 85,000 M−1cm−1.

Acridine orange is an organic compound that serves as a nucleic acid-selective fluorescent dye with cationic properties useful for cell cycle determination. Acridine orange is cell-permeable, which allows the dye to interact with DNA by intercalation, or RNA via electrostatic attractions. When bound to DNA, acridine orange is very similar spectrally to an organic compound known as fluorescein. Acridine orange and fluorescein have a maximum excitation at 502nm and 525 nm (green). When acridine orange associates with RNA, the fluorescent dye experiences a maximum excitation shift from 525 nm (green) to 460 nm (blue). The shift in maximum excitation also produces a maximum emission of 650 nm (red). Acridine orange is able to withstand low pH environments, allowing the fluorescent dye to penetrate acidic organelles such as lysosomes and phagolysosomes that are membrane-bound organelles essential for acid hydrolysis or for producing products of phagocytosis of apoptotic cells. Acridine orange is used in epifluorescence microscopy and flow cytometry. The ability to penetrate the cell membranes of acidic organelles and cationic properties of acridine orange allows the dye to differentiate between various types of cells. The shift in maximum excitation and emission wavelengths provides a foundation to predict the wavelength at which the cells will stain.

Rhodamine 123 is a chemical compound and a dye. It is often used as a tracer dye within water to determine the rate and direction of flow and transport. Rhodamine dyes fluoresce and can thus be detected easily and inexpensively with instruments called fluorometers. Rhodamine dyes are used extensively in biotechnology applications such as fluorescence microscopy, flow cytometry, fluorescence correlation spectroscopy and ELISA. Rhodamine fluorescence can also be used as a measure of membrane polarization in live cell assays both within mitochondria and with bacteria. This use relies on the fact that rhodamine 123 accumulates in membranes in a manner which is dependent on membrane polarization.

Fluorescence is used in the life sciences generally as a non-destructive way of tracking or analysing biological molecules. Some proteins or small molecules in cells are naturally fluorescent, which is called intrinsic fluorescence or autofluorescence. Alternatively, specific or general proteins, nucleic acids, lipids or small molecules can be "labelled" with an extrinsic fluorophore, a fluorescent dye which can be a small molecule, protein or quantum dot. Several techniques exist to exploit additional properties of fluorophores, such as fluorescence resonance energy transfer, where the energy is passed non-radiatively to a particular neighbouring dye, allowing proximity or protein activation to be detected; another is the change in properties, such as intensity, of certain dyes depending on their environment allowing their use in structural studies.
Microfluorimetry is an adaption of fluorimetry for studying the biochemical and biophysical properties of cells by using microscopy to image cell components tagged with fluorescent molecules. It is a type of microphotometry that gives a quantitative measure of the qualitative nature of fluorescent measurement and therefore, allows for definitive results that would have been previously indiscernible to the naked eye.
Super-resolution microscopy is a series of techniques in optical microscopy that allow such images to have resolutions higher than those imposed by the diffraction limit, which is due to the diffraction of light. Super-resolution imaging techniques rely on the near-field or on the far-field. Among techniques that rely on the latter are those that improve the resolution only modestly beyond the diffraction-limit, such as confocal microscopy with closed pinhole or aided by computational methods such as deconvolution or detector-based pixel reassignment, the 4Pi microscope, and structured-illumination microscopy technologies such as SIM and SMI.
Chromatin bridge is a mitotic occurrence that forms when telomeres of sister chromatids fuse together and fail to completely segregate into their respective daughter cells. Because this event is most prevalent during anaphase, the term anaphase bridge is often used as a substitute. After the formation of individual daughter cells, the DNA bridge connecting homologous chromosomes remains fixed. As the daughter cells exit mitosis and re-enter interphase, the chromatin bridge becomes known as an interphase bridge. These phenomena are usually visualized using the laboratory techniques of staining and fluorescence microscopy.
Photo-activated localization microscopy and stochastic optical reconstruction microscopy (STORM) are widefield fluorescence microscopy imaging methods that allow obtaining images with a resolution beyond the diffraction limit. The methods were proposed in 2006 in the wake of a general emergence of optical super-resolution microscopy methods, and were featured as Methods of the Year for 2008 by the Nature Methods journal. The development of PALM as a targeted biophysical imaging method was largely prompted by the discovery of new species and the engineering of mutants of fluorescent proteins displaying a controllable photochromism, such as photo-activatible GFP. However, the concomitant development of STORM, sharing the same fundamental principle, originally made use of paired cyanine dyes. One molecule of the pair, when excited near its absorption maximum, serves to reactivate the other molecule to the fluorescent state.

Methyl green is a cationic or positive charged stain related to Ethyl Green that has been used for staining DNA since the 19th century. It has been used for staining cell nuclei either as a part of the classical Unna-Pappenheim stain or as a nuclear counterstain ever since.
In recent years, its fluorescent properties, when bound to DNA, have positioned it as useful for far-red imaging of live cell nuclei. Fluorescent DNA staining is routinely used in cancer prognosis. Methyl green also emerges as an alternative stain for DNA in agarose gels, fluorometric assays, and flow cytometry. It has also been shown that it can be used as an exclusion viability stain for cells. Its interaction with DNA has been shown to be non-intercalating, in other words, not inserting itself into the DNA, but instead electrostatic with the DNA major groove. It is used in combination with pyronin in the methyl green–pyronin stain, which stains and differentiates DNA and RNA.
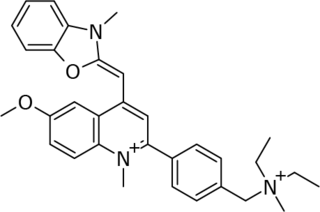
SYBR Gold is an asymmetrical cyanine dye. It can be used as a stain for double-stranded DNA, single-stranded DNA, and RNA. SYBR Gold is the most sensitive fluorescent stain of the SYBR family of dyes for the detection of nucleic acids. The SYBR family of dyes is produced by Molecular Probes Inc., now owned by Thermo Fisher Scientific