Nitrogen fixation is a chemical process by which molecular dinitrogen is converted into ammonia. It occurs both biologically and abiologically in chemical industries. Biological nitrogen fixation or diazotrophy is catalyzed by enzymes called nitrogenases. These enzyme complexes are encoded by the Nif genes and contain iron, often with a second metal.

Heterocysts or heterocytes are specialized nitrogen-fixing cells formed during nitrogen starvation by some filamentous cyanobacteria, such as Nostoc, Cylindrospermum, and Anabaena. They fix nitrogen from dinitrogen (N2) in the air using the enzyme nitrogenase, in order to provide the cells in the filament with nitrogen for biosynthesis.
Diazotrophs are bacteria and archaea that fix atmospheric nitrogen (N2) in the atmosphere into bioavailable forms such as ammonia.
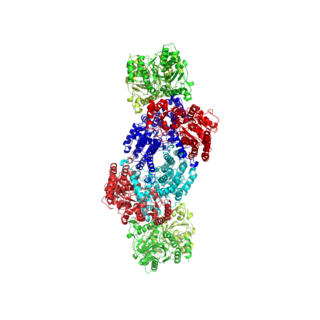
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

The Pseudomonadaceae are a family of bacteria which includes the genera Azomonas, Azorhizophilus, Azotobacter, Mesophilobacter, Pseudomonas, and Rugamonas. The family Azotobacteraceae was recently reclassified into this family.

Azotobacter is a genus of usually motile, oval or spherical bacteria that form thick-walled cysts and may produce large quantities of capsular slime. They are aerobic, free-living soil microbes that play an important role in the nitrogen cycle in nature, binding atmospheric nitrogen, which is inaccessible to plants, and releasing it in the form of ammonium ions into the soil. In addition to being a model organism for studying diazotrophs, it is used by humans for the production of biofertilizers, food additives, and some biopolymers. The first representative of the genus, Azotobacter chroococcum, was discovered and described in 1901 by Dutch microbiologist and botanist Martinus Beijerinck. Azotobacter species are Gram-negative bacteria found in neutral and alkaline soils, in water, and in association with some plants.
Glutamine synthetase (GS) is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:

Rhodospirillum rubrum is a Gram-negative, pink-coloured bacterium, with a size of 0.8-1 µm. It is a facultative anaerobe with a large set of possible metabolisms depending on the conditions of its environment. It's capable of using oxygen for aerobic respiration under aerobic conditions, or an alternative terminal electron acceptor for anaerobic respiration under anaerobic conditions. Alternative terminal electron acceptors for R. rubrum include dimethyl sulfoxide or trimethylamine oxide.
Carbamoyl phosphate synthetase I is a ligase enzyme located in the mitochondria involved in the production of urea. Carbamoyl phosphate synthetase I transfers an ammonia molecule to a molecule of bicarbonate that has been phosphorylated by a molecule of ATP. The resulting carbamate is then phosphorylated with another molecule of ATP. The resulting molecule of carbamoyl phosphate leaves the enzyme.

Cobalt chelatase (EC 6.6.1.2) is an enzyme that catalyzes the chemical reaction

Vanadium nitrogenase is a key enzyme for nitrogen fixation found in nitrogen-fixing bacteria, and is used as an alternative to molybdenum nitrogenase when molybdenum is unavailable. Vanadium nitrogenases are an important biological use of vanadium, which is uncommonly used by life. An important component of the nitrogen cycle, vanadium nitrogenase converts nitrogen gas to ammonia, thereby making otherwise inaccessible nitrogen available to plants. Unlike molybdenum nitrogenase, vanadium nitrogenase can also reduce carbon monoxide to ethylene, ethane and propane but both enzymes can reduce protons to hydrogen gas and acetylene to ethylene.

The Nif regulon is a set of seven operons used to regulate nitrogen fixation in the coliform bacterium Klebsiella pneumoniae under anaerobic and microaerophilic conditions. It includes 17 nif genes, and is situated between the his and the Shi-A operon of the bacterium.

The PII family comprises a group of widely distributed signal transduction proteins found in nearly all Bacteria and also present in Archaea and in the chloroplasts of Algae and plants. PII form barrel-like homotrimers with a flexible loop, namely T-loop, emerging from each subunit. PII proteins have extraordinary sensory properties; they can exist in a vast range of structural status accordingly to the levels of ATP, ADP and 2-oxogluratate. These metabolites interact allosterically with PII in three conserved binding sites located in the lateral cavity between each PII subunit. ATP and ADP bind competitively to the nucleotide binding whereas the 2-oxoglutarate only interacts with PII in the presence of MgATP.

Methanococcus maripaludis is a species of methanogenic archaea found in marine environments, predominantly salt marshes. M. maripaludis is a non-pathogenic, gram-negative, weakly motile, non-spore-forming, and strictly anaerobic mesophile. It is classified as a chemolithoautotroph. This archaeon has a pleomorphic coccoid-rod shape of 1.2 by 1.6 μm, in average size, and has many unique metabolic processes that aid in survival. M. maripaludis also has a sequenced genome consisting of around 1.7 Mbp with over 1,700 identified protein-coding genes. In ideal conditions, M. maripaludis grows quickly and can double every two hours.
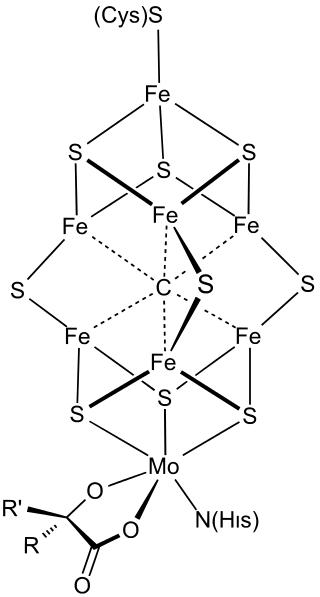
FeMoco (FeMo cofactor) is the primary cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Because it contains iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C.

Cyanothece is a genus of unicellular, diazotrophic, oxygenic photosynthesizing cyanobacteria.

Azotobacter chroococcum is a bacterium that has the ability to fix atmospheric nitrogen. It was discovered by Martinus Beijerinck in 1901, and was the first aerobic, free-living nitrogen fixer discovered. A. chroococcum could be useful for nitrogen fixation in crops as a biofertilizer, fungicide, and nutrient indicator, and in bioremediation.
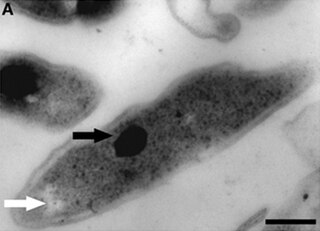
Methylacidiphilum fumariolicum is an autotrophic bacterium first described in 2007 growing on volcanic pools near Naples, Italy. It grows in mud at temperatures between 50 °C and 60 °C and an acidic pH of 2–5. It is able to oxidize methane gas. It uses ammonium, nitrate or atmospheric nitrogen as a nitrogen source and fixes carbon dioxide.
Douglas Charles "Doug" Rees is an American biochemist, biophysicist, and structural biologist.













