Related Research Articles
An integrated development environment (IDE) is a software application that provides comprehensive facilities for software development. An IDE normally consists of at least a source-code editor, build automation tools, and a debugger. Some IDEs, such as IntelliJ IDEA, NetBeans, and Eclipse, contain the necessary compiler, interpreter, or both; others, such as SharpDevelop and Lazarus, do not.

Structural biology is a field that is many centuries old which, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization. Early structural biologists throughout the 19th and early 20th centuries were primarily only able to study structures to the limit of the naked eye's visual acuity and through magnifying glasses and light microscopes.

QNX is a commercial Unix-like real-time operating system, aimed primarily at the embedded systems market.

wxWidgets is a widget toolkit and tools library for creating graphical user interfaces (GUIs) for cross-platform applications. wxWidgets enables a program's GUI code to compile and run on several computer platforms with minimal or no code changes. A wide choice of compilers and other tools to use with wxWidgets facilitates development of sophisticated applications. wxWidgets supports a comprehensive range of popular operating systems and graphical libraries, both proprietary and free, and is widely deployed in prominent organizations.
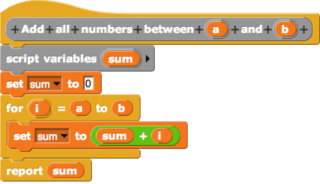
In computing, a visual programming language or block coding is a programming language that lets users create programs by manipulating program elements graphically rather than by specifying them textually. A VPL allows programming with visual expressions, spatial arrangements of text and graphic symbols, used either as elements of syntax or secondary notation. For example, many VPLs are based on the idea of "boxes and arrows", where boxes or other screen objects are treated as entities, connected by arrows, lines or arcs which represent relations.

Geographic Resources Analysis Support System is a geographic information system (GIS) software suite used for geospatial data management and analysis, image processing, producing graphics and maps, spatial and temporal modeling, and visualizing. It can handle raster, topological vector, image processing, and graphic data.
JMP is a suite of computer programs for statistical analysis developed by JMP, a subsidiary of SAS Institute. It was launched in 1989 to take advantage of the graphical user interface introduced by the Macintosh operating systems. It has since been significantly rewritten and made available also for the Windows operating system. JMP is used in applications such as Six Sigma, quality control, and engineering, design of experiments, as well as for research in science, engineering, and social sciences.
Molecular graphics is the discipline and philosophy of studying molecules and their properties through graphical representation. IUPAC limits the definition to representations on a "graphical display device". Ever since Dalton's atoms and Kekulé's benzene, there has been a rich history of hand-drawn atoms and molecules, and these representations have had an important influence on modern molecular graphics.
The CCP4 file format is file generated by the Collaborative Computational Project Number 4 in 1979. The file format for electron density has become industry standard in X-ray crystallography and Cryo-electron microscopy where the result of the technique is a three-dimensional grid of voxels each with a value corresponding to density of electrons The CCP4 format is supported by almost every molecular graphics suite that supports volumetric data. The major packages include:

BALL is a C++ class framework and set of algorithms and data structures for molecular modelling and computational structural bioinformatics, a Python interface to this library, and a graphical user interface to BALL, the molecule viewer BALLView.
Foldit is an online puzzle video game about protein folding. It is part of an experimental research project developed by the University of Washington, Center for Game Science, in collaboration with the UW Department of Biochemistry. The objective of Foldit is to fold the structures of selected proteins as perfectly as possible, using tools provided in the game. The highest scoring solutions are analyzed by researchers, who determine whether or not there is a native structural configuration that can be applied to relevant proteins in the real world. Scientists can then use these solutions to target and eradicate diseases and create biological innovations. A 2010 paper in the science journal Nature credited Foldit's 57,000 players with providing useful results that matched or outperformed algorithmically computed solutions.

The program Coot is used to display and manipulate atomic models of macromolecules, typically of proteins or nucleic acids, using 3D computer graphics. It is primarily focused on building and validation of atomic models into three-dimensional electron density maps obtained by X-ray crystallography methods, although it has also been applied to data from electron microscopy.

The Environmental Molecular Sciences Laboratory is a Department of Energy, Office of Science facility at Pacific Northwest National Laboratory in Richland, Washington, United States.

The Collaborative Computing Project for NMR (CCPN) is a project that aims to bring together computational aspects of the scientific community involved in NMR spectroscopy, especially those who work in the field of protein NMR. The general aims are to link new and existing NMR software via a common data standard and provide a forum within the community for the discussion of NMR software and the scientific methods it supports. CCPN was initially started in 1999 in the United Kingdom but collaborates with NMR and software development groups worldwide.
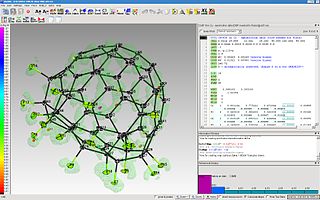
The program ShelXle is a graphical user interface for the structure refinement program SHELXL. ShelXle combines an editor with syntax highlighting for the SHELXL-associated .ins (input) and .res (output) files with an interactive graphical display for visualization of a three-dimensional structure including the electron density (Fo) and difference density (Fo-Fc) maps.
Scigress, stylized SCiGRESS, is a software suite for molecular modelling, computational chemistry, drug design, and materials science, a successor to Computer Aided Chemistry (CAChe) software.

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.

Paul Emsley is a British crystallographer at the MRC Laboratory of Molecular Biology in Cambridge. He works as an independent scientist and is a member of the Computational Crystallography Group headed by Garib Murshudov.
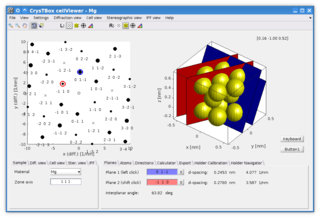
CrysTBox is a suite of computer tools designed to accelerate material research based on transmission electron microscope images via highly accurate automated analysis and interactive visualization. Relying on artificial intelligence and computer vision, CrysTBox makes routine crystallographic analyses simpler, faster and more accurate compared to human evaluators. The high level of automation together with sub-pixel precision and interactive visualization makes the quantitative crystallographic analysis accessible even for non-crystallographers allowing for an interdisciplinary research. Simultaneously, experienced material scientists can take advantage of advanced functionalities for comprehensive analyses.
References
- ↑ M.D. Winn, C.C. Ballard, K.D. Cowtan, E.J. Dodson, P. Emsley, P.R. Evans, R.M. Keegan, E.B. Krissinel, A.G.W. Leslie, A. McCoy, S.J. McNicholas, G.N. Murshudov, N.S. Pannu, E.A. Potterton, H.R. Powell, R.J. Read, A. Vagin, K.S. Wilson (2011) Overview of the CCP4 suite and current developments Acta Crystallogr. D67, 235-242