Related Research Articles

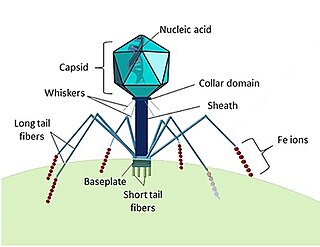
A bacteriophage, also known informally as a phage, is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

Cyanobacteria, also known as Cyanophyta, are a phylum of Gram-negative bacteria that obtain energy via photosynthesis. The name cyanobacteria refers to their color, which similarly forms the basis of cyanobacteria's common name, blue-green algae. They appear to have originated in a freshwater or terrestrial environment. Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.

Filamentous bacteriophage is a family of viruses (Inoviridae) that infect bacteria. The phages are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. The coat of the virion comprises five types of viral protein, which are located during phage assembly in the inner membrane of the host bacteria, and are added to the nascent virion as it extrudes through the membrane. The simplicity of this family makes it an attractive model system to study fundamental aspects of molecular biology, and it has also proven useful as a tool in immunology and nanotechnology.

An arenavirus is a bisegmented ambisense RNA virus that is a member of the family Arenaviridae. These viruses infect rodents and occasionally humans. A class of novel, highly divergent arenaviruses, properly known as reptarenaviruses, have also been discovered which infect snakes to produce inclusion body disease. At least eight arenaviruses are known to cause human disease. The diseases derived from arenaviruses range in severity. Aseptic meningitis, a severe human disease that causes inflammation covering the brain and spinal cord, can arise from the lymphocytic choriomeningitis virus. Hemorrhagic fever syndromes, including Lassa fever, are derived from infections such as Guanarito virus, Junin virus, Lassa virus, Lujo virus, Machupo virus, Sabia virus, or Whitewater Arroyo virus. Because of the epidemiological association with rodents, some arenaviruses and bunyaviruses are designated as roboviruses.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.

Cyanophages are viruses that infect cyanobacteria, also known as Cyanophyta or blue-green algae. Cyanobacteria are a phylum of bacteria that obtain their energy through the process of photosynthesis. Although cyanobacteria metabolize photoautotrophically like eukaryotic plants, they have prokaryotic cell structure. Cyanophages can be found in both freshwater and marine environments. Marine and freshwater cyanophages have icosahedral heads, which contain double-stranded DNA, attached to a tail by connector proteins. The size of the head and tail vary among species of cyanophages. Cyanophages infect a wide range of cyanobacteria and are key regulators of the cyanobacterial populations in aquatic environments, and may aid in the prevention of cyanobacterial blooms in freshwater and marine ecosystems. These blooms can pose a danger to humans and other animals, particularly in eutrophic freshwater lakes. Infection by these viruses is highly prevalent in cells belonging to Synechococcus spp. in marine environments, where up to 5% of cells belonging to marine cyanobacterial cells have been reported to contain mature phage particles.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
Rice hoja blanca tenuivirus (RHBV), meaning "white leaf rice virus", is a plant virus in the family Phenuiviridae. RHBV causes Hoja blanca disease (HBD), which affects the leaves of the rice plant Oryza sativa, stunting the growth of the plant or killing it altogether. RHBV is carried by an insect vector, Tagosodes orizicolus, a type of planthopper. The virus is found in South America, Mexico, throughout Central America, the Caribbean region, and the southern United States. In South America, the disease is endemic to Colombia, Venezuela, Ecuador, Peru, Suriname, French Guiana and Guyana.
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip.

Corticovirus is a genus of viruses in the family Corticoviridae. Corticoviruses are bacteriophages; that is, their natural hosts are bacteria. The genus contains two species. The name is derived from Latin cortex, corticis. However, prophages closely related to PM2 are abundant in the genomes of aquatic bacteria, suggesting that the ecological importance of corticoviruses might be underestimated. Bacteriophage PM2 was first described in 1968 after isolation from seawater sampled from the coast of Chile.
Cyanobionts are cyanobacteria that live in symbiosis with a wide range of organisms such as terrestrial or aquatic plants; as well as, algal and fungal species. They can reside within extracellular or intracellular structures of the host. In order for a cyanobacterium to successfully form a symbiotic relationship, it must be able to exchange signals with the host, overcome defense mounted by the host, be capable of hormogonia formation, chemotaxis, heterocyst formation, as well as possess adequate resilience to reside in host tissue which may present extreme conditions, such as low oxygen levels, and/or acidic mucilage. The most well-known plant-associated cyanobionts belong to the genus Nostoc. With the ability to differentiate into several cell types that have various functions, members of the genus Nostoc have the morphological plasticity, flexibility and adaptability to adjust to a wide range of environmental conditions, contributing to its high capacity to form symbiotic relationships with other organisms. Several cyanobionts involved with fungi and marine organisms also belong to the genera Richelia, Calothrix, Synechocystis, Aphanocapsa and Anabaena, as well as the species Oscillatoria spongeliae. Although there are many documented symbioses between cyanobacteria and marine organisms, little is known about the nature of many of these symbioses. The possibility of discovering more novel symbiotic relationships is apparent from preliminary microscopic observations.
A viral structural protein is a viral protein that is a structural component of the mature virus.

Escherichia virus T5, sometimes called Bacteriophage T5 is a caudal virus within the family Demerecviridae. This bacteriophage specifically infects E. coli bacterial cells and follows a lytic life cycle.
Phikmvvirus is a genus of viruses that infect bacteria. There are currently 16 species in this genus including the type species Pseudomonas virus phiKMV. Bacteriophage phiKMV and its relatives are known to be highly virulent phages, producing large clear plaques on a susceptible host. The only reported exception is phage LKA1, which yields small plaques surrounded by a halo. While all other P. aeruginosa-specific phikmvviruses use the Type IV pili as primary receptor, LKA1 particles attach to the bacterial lipopolysaccharide layer.

Ff phages is a group of almost identical filamentous phage including phages f1, fd, M13 and ZJ/2, which infect bacteria bearing the F fertility factor. The virion is a flexible filament measuring about 6 by 900 nm, comprising a cylindrical protein tube protecting a single-stranded circular DNA molecule at its core. The phage codes for only 11 gene products, and is one of the simplest viruses known. It has been widely used to study fundamental aspects of molecular biology. George Smith and Greg Winter used f1 and fd for their work on phage display for which they were awarded a share of the 2018 Nobel Prize in Chemistry. Early experiments on Ff phages used M13 to identify gene functions, and M13 was also developed as a cloning vehicle, so the name M13 is sometimes used as an informal synonym for the whole group of Ff phages.
Tristromaviridae is a family of viruses. Archaea of the genera Thermoproteus and Pyrobaculum serve as natural hosts. Tristromaviridae is the sole family in the order Primavirales. There are two genera and three species in the family.
Auxiliary metabolic genes (AMGs) are found in many bacteriophages but originated in bacterial cells. AMGs modulate host cell metabolism during infection so that the phage can replicate more efficiently. For instance, bacteriophages that infect the abundant marine cyanobacteria Synechococcus and Prochlorococcus (cyanophages) carry AMGs that have been acquired from their immediate host as well as more distantly-related bacteria. Cyanophage AMGs support a variety of functions including photosynthesis, carbon metabolism, nucleic acid synthesis and metabolism.

Pseudomonas virus gh1 is a bacteriophage capable of infecting susceptible strains of Pseudomonas putida. It is a member of family Podoviridae, subfamily Autographivirinae. It was first isolated in 1966 from a sample taken from the aeration tank at a sewage plant in East Lansing, Michigan.
Marine viruses are defined by their habitat as viruses that are found in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Viruses are small infectious agents that can only replicate inside the living cells of a host organism, because they need the replication machinery of the host to do so. They can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.
References
- 1 2 3 Chénard C, Wirth JF, Suttle CA (2016), "Viruses Infecting a freshwater filamentous cyanobacterium (Nostoc sp.) encode a functional CRISPR array and a proteobacterial DNA polymerase B", mBio , 7: e00667-16, doi:10.1128/mBio.00667-16, PMC 4916379 , PMID 27302758
- 1 2 Adolph KW, Haselkorn R (1971), "Isolation and characterization of a virus infecting the blue-green alga Nostoc muscorum", Virology , 46: 200–208, doi:10.1016/0042-6822(71)90023-7
- ↑ Sarma 2012, pp. 420, 423
- ↑ Sarma 2012, p. 423
- ↑ Adolph KW, Haselkorn R (1973), "Blue-green algal virus N-1: Physical properties and disassembly into structural parts", Virology , 53: 427–40, doi:10.1016/0042-6822(73)90222-5
- ↑ Sarma 2012, pp. 427, 429
- ↑ Sarma 2012, p. 430
- ↑ Calendar 2006, p. 524
- ↑ Sarma 2012, p. 433
Sources
- Calendar R. The Bacteriophages (Oxford University Press; 2006) ( ISBN 0195148509)
- Sarma TA. Handbook of Cyanobacteria (CRC Press; 2012) ( ISBN 1466559411)