Related Research Articles

Lactococcus lactis is a gram-positive bacterium used extensively in the production of buttermilk and cheese, but has also become famous as the first genetically modified organism to be used alive for the treatment of human disease. L. lactis cells are cocci that group in pairs and short chains, and, depending on growth conditions, appear ovoid with a typical length of 0.5 - 1.5 μm. L. lactis does not produce spores (nonsporulating) and are not motile (nonmotile). They have a homofermentative metabolism, meaning they produce lactic acid from sugars. They've also been reported to produce exclusive L-(+)-lactic acid. However, reported D-(−)-lactic acid can be produced when cultured at low pH. The capability to produce lactic acid is one of the reasons why L. lactis is one of the most important microorganisms in the dairy industry. Based on its history in food fermentation, L. lactis has generally recognized as safe (GRAS) status, with few case reports of it being an opportunistic pathogen.

Acidobacteriota is a phylum of Gram-negative bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.

Clostridium acetobutylicum, ATCC 824, is a commercially valuable bacterium sometimes called the "Weizmann Organism", after Jewish Russian-born biochemist Chaim Weizmann. A senior lecturer at the University of Manchester, England, he used them in 1916 as a bio-chemical tool to produce at the same time, jointly, acetone, ethanol, and n-butanol from starch. The method has been described since as the ABE process,, yielding 3 parts of acetone, 6 of n-butanol, and 1 of ethanol. Acetone was used in the important wartime task of casting cordite. The alcohols were used to produce vehicle fuels and synthetic rubber.
Desulfovibrio is a genus of Gram-negative sulfate-reducing bacteria. Desulfovibrio species are commonly found in aquatic environments with high levels of organic material, as well as in water-logged soils, and form major community members of extreme oligotrophic habitats such as deep granitic fractured rock aquifers. They're also found in the guts of beetles, such as Melolontha melolontha, where they perform sulfate reduction.
Ultramicrobacteria are bacteria that are smaller than 0.1 μm3 under all growth conditions. This term was coined in 1981, describing cocci in seawater that were less than 0.3 μm in diameter. Ultramicrobacteria have also been recovered from soil and appear to be a mixture of gram-positive, gram-negative and cell-wall-lacking species. Ultramicrobacteria possess a relatively high surface-area-to-volume ratio due to their small size, which aids in growth under oligotrophic conditions. The relatively small size of ultramicrobacteria also enables parasitism of larger organisms; some ultramicrobacteria have been observed to be obligate or facultative parasites of various eukaryotes and prokaryotes. One factor allowing ultramicrobacteria to achieve their small size seems to be genome minimization such as in the case of the ultramicrobacterium P. ubique whose small 1.3 Mb genome is seemingly devoid of extraneous genetic elements like non-coding DNA, transposons, extrachromosomal elements etc. However, genomic data from ultramicrobacteria is lacking since the study of ultramicrobacteria, like many other prokaryotes, is hindered by difficulties in cultivating them.

L-form bacteria, also known as L-phase bacteria, L-phase variants or cell wall-deficient bacteria (CWDB), are growth forms derived from different bacteria. They lack cell walls. Two types of L-forms are distinguished: unstable L-forms, spheroplasts that are capable of dividing, but can revert to the original morphology, and stable L-forms, L-forms that are unable to revert to the original bacteria.
Cupriavidus metallidurans is a non-spore-forming, Gram-negative bacterium which is adapted to survive several forms of heavy metal stress.

Nitratidesulfovibrio vulgaris is a species of Gram-negative sulfate-reducing bacteria in the Desulfovibrionaceae family. It is also an anaerobic sulfate-reducing bacterium that is an important organism involved in the bioremediation of heavy metals in the environment. Nitratidesulfovibrio vulgaris is often used as a model organism for sulfur-reducing bacteria and was the first of such bacteria to have its genome sequenced. It is ubiquitous in nature and has also been implicated in a variety of human bacterial infections, although it may only be an opportunistic pathogen. This microbe also has the ability to endure high salinity environments, which is done through the utilization of osmoprotectants and efflux systems.
Elusimicrobium minutum is an ultramicrobacterium and first accepted member to be cultured of a major bacterial lineage previously known only as candidate phylum Termite Gut 1 (TG1), which has accordingly been renamed phylum Elusimicrobiota. It was isolated in the laboratory of Andreas Brune at the Max Planck Institute for Terrestrial Microbiology, from the scarab beetle. It is a mesophilic, obligately anaerobic ultramicrobacterium with a gram-negative cell envelope. Cells are typically rod shaped, but cultures are pleomorphic in all growth phases. The isolate grows heterotrophically on sugars and ferments D-galactose, D-glucose, D-fructose, D-glucosamine, and N-acetyl-D-glucosamine to acetate, ethanol, hydrogen, and alanine as major products but only if amino acids are present in the medium
The phylum Elusimicrobiota, previously known as "Termite Group 1", has been shown to be widespread in different ecosystems like marine environment, sewage sludge, contaminated sites and soils, and toxic wastes. The high abundance of Elusimicrobiota representatives is only seen for the lineage of symbionts found in termites and ants.
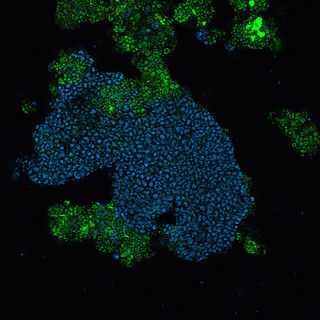
Candidatus Accumulibacter phosphatis (CAP) is an unclassified type of Betaproteobacteria that is a common bacterial community member of sewage treatment and wastewater treatment plants performing enhanced biological phosphorus removal (EBPR) and is a polyphosphate-accumulating organism. The role of CAP in EBPR was elucidated using culture-independent approaches such as 16S rRNA clone banks that showed the Betaproteobacteria dominated lab-scale EBPR reactors. Further work using clone banks and fluorescence in situ hybridization identified a group of bacteria, closely related to Rhodocyclus as the dominant member of lab-scale communities.
Syntrophobacter wolinii is a non-motile, gram-negative and rod-shaped species of bacteria that was originally isolated from a wastewater digester. This species is able to perform propionate degradation and sulfate reduction. S. wolinii can be grown in co-culture or pure culture. 16s rRNA analysis shows its close relation to other sulfate reducers.
Dehalogenimonas lykanthroporepellens is an anaerobic, Gram-negative bacteria in the phylum Chloroflexota isolated from a Superfund site in Baton Rouge, Louisiana. It is useful in bioremediation for its ability to reductively dehalogenate chlorinated alkanes.
Methanosaeta concilii is an archaeum in the disputed genus Methanosaeta. It is obligately anaerobic, gram-negative and non-motile. It is rod-shaped with flat ends. The cells are enclosed within a cross-striated sheath. The type strain is GP6. Its genome has been sequenced.
Desulfocapsa thiozymogenes is an anaerobic, gram-negative bacterium. It disproportionates elemental sulfur. It is the type species of its genus.
Congregibacter litoralis KT71 is a gram-negative Gammaproteobacteria part of the NOR5/OM60 Clade discovered in seawater from Heligoland, an island in the North Sea by H. Eilers from the Max Planck Institute for Microbiology. C. litoralis KT71 is described as a pleomorphic bacterium and has a size of 2 x 0.5 μm. When grown in culture, C. litoralis KT71 has a generation time of 4.5 hours and prefers to grow on complex substrates where the sole carbon source is undefined, though it can utilize some sole carbon sources because they are most likely used by the organism for its central metabolism.
Haladaptatus paucihalophilus is a halophilic archaeal species, originally isolated from a spring in Oklahoma. It uses a new pathway to synthesize glycine, and contains unique physiological features for osmoadaptation.
Dokdonia donghaensis is a strictly aerobic, gram-negative, phototrophic bacterium that thrives in marine environments. The organism can grow at a broad range of temperatures on seawater media. It has the ability to form biofilms, which increases the organism's resistance to antimicrobial agents, such as tetracycline.
Mercury methylation is the process of forming methylmercury (MeHg). The methylation of mercury can occur abiotically or biotically. Biotically, the primary methylators of mercury are sulfate-reducing and iron-reducing bacteria. Three mechanisms have been proposed for the biotic methylation of mercury by sulfate-reducing bacteria. Mercury methylation can be problematic as methylmercury is toxic and can be bio-magnified through the food web.
References
- ↑ Jean, Euzeby. "Genus Desulfovibrio". List of Prokaryotic names with Standing in Nomenclature . Retrieved November 6, 2014.
- ↑ EJC Goldstein; DM Citron; VA Peraino; SA Cross (June 2003). "Desulfovibrio desulfricans Bacteremia and Review of Human Desulfovibrio Infections". J Clin Microbiol. 41 (6): 2752–2754. doi:10.1128/jcm.41.6.2752-2754.2003. PMC 156571 . PMID 12791922.
- ↑ CC Gilmour; DA Elias; AM Kucken; SD Brown; AV Palumbo; CW Schadt; JD Wall (June 2011). "Sulfate-Reducing Bacterium Desulfovibrio desulfuricans ND132 as a Model for Understanding Bacterial Mercury Methylation". Appl Environ Microbiol. 77 (12): 3938–3951. Bibcode:2011ApEnM..77.3938G. doi:10.1128/aem.02993-10. PMC 3131654 . PMID 21515733.
- ↑ Sánchez-Andrea, I., Guedes, I.A., Hornung, B. et al. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat Commun 11, 5090 (2020). https://doi.org/10.1038/s41467-020-18906-7
- ↑ "Home - Desulfovibrio desulfuricans G20". genome.jgi.doe.gov. Retrieved 2017-05-07.
- ↑ Rapp, Barbara J., and Judy D. Wall. "Genetic transfer in Desulfovibrio desulfuricans." Proceedings of the National Academy of Sciences 84.24 (1987): 9128-9130.
- ↑ GC Compeau; R Bartha (August 1985). "Sulfate-Reducing Bacteria: Principal Methylators of Mercury in Anoxic Estuarine Sediment". Appl Environ Microbiol. 50 (2): 498–502. Bibcode:1985ApEnM..50..498C. doi:10.1128/AEM.50.2.498-502.1985. PMC 238649 . PMID 16346866.