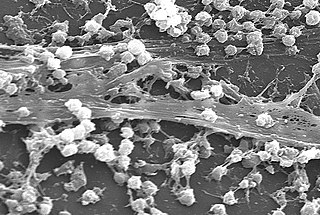
A biofilm comprises any syntrophic consortium of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric conglomeration of extracellular polysaccharides, proteins, lipids and DNA. Because they have three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes".

Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Listeria monocytogenes is the species of pathogenic bacteria that causes the infection listeriosis. It is a facultative anaerobic bacterium, capable of surviving in the presence or absence of oxygen. It can grow and reproduce inside the host's cells and is one of the most virulent foodborne pathogens: 20 to 30% of foodborne listeriosis infections in high-risk individuals may be fatal. In the European Union, listeriosis follows an upward trend that began in 2008, causing 2,161 confirmed cases and 210 reported deaths in 2014, 16% more than in 2013. Listeriosis mortality rates are also higher in the EU than for other foodborne pathogens. Responsible for an estimated 1,600 illnesses and 260 deaths in the United States annually, listeriosis ranks third in total number of deaths among foodborne bacterial pathogens, with fatality rates exceeding even Salmonella spp. and Clostridium botulinum.

Proteus is a genus of Gram-negative bacteria. It is a rod shaped, aerobic and motile bacteria, which is able to migrate across surfaces due its “swarming” characteristic in temperatures between 20 and 37 °C. Their size generally ranges from 0.4–0.8 μm in diameter and 1.0–3.0 μm in length. They tend to have an ammonia smell. Proteus bacilli are widely distributed in nature as saprophytes, being found in decomposing animal matter, sewage, manure soil, the mammalian intestine, and human and animal feces. They are opportunistic pathogens, commonly responsible for urinary and septic infections, often nosocomial.
A slime layer in bacteria is an easily removable, unorganized layer of extracellular material that surrounds bacteria cells. Specifically, this consists mostly of exopolysaccharides, glycoproteins, and glycolipids. Therefore, the slime layer is considered as a subset of glycocalyx.

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.
Adhesins are cell-surface components or appendages of bacteria that facilitate adhesion or adherence to other cells or to surfaces, usually in the host they are infecting or living in. Adhesins are a type of virulence factor.

Staphylococcus epidermidis is a Gram-positive bacterium, and one of over 40 species belonging to the genus Staphylococcus. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota and also found in marine sponges. It is a facultative anaerobic bacteria. Although S. epidermidis is not usually pathogenic, patients with compromised immune systems are at risk of developing infection. These infections are generally hospital-acquired. S. epidermidis is a particular concern for people with catheters or other surgical implants because it is known to form biofilms that grow on these devices. Being part of the normal skin microbiota, S. epidermidis is a frequent contaminant of specimens sent to the diagnostic laboratory.

Shigella flexneri is a species of Gram-negative bacteria in the genus Shigella that can cause diarrhea in humans. Several different serogroups of Shigella are described; S. flexneri belongs to group B. S. flexneri infections can usually be treated with antibiotics, although some strains have become resistant. Less severe cases are not usually treated because they become more resistant in the future. Shigella are closely related to Escherichia coli, but can be differentiated from E.coli based on pathogenicity, physiology and serology.
Pantoea agglomerans is a Gram-negative bacterium that belongs to the family Erwiniaceae.

Shewanella oneidensis is a bacterium notable for its ability to reduce metal ions and live in environments with or without oxygen. This proteobacterium was first isolated from Lake Oneida, NY in 1988, hence its name.
Osmoprotectants or compatible solutes are small organic molecules with neutral charge and low toxicity at high concentrations that act as osmolytes and help organisms survive extreme osmotic stress. Osmoprotectants can be placed in three chemical classes: betaines and associated molecules, sugars and polyols, and amino acids. These molecules accumulate in cells and balance the osmotic difference between the cell's surroundings and the cytosol. In plants, their accumulation can increase survival during stresses such as drought. In extreme cases, such as in bdelloid rotifers, tardigrades, brine shrimp, and nematodes, these molecules can allow cells to survive being completely dried out and let them enter a state of suspended animation called cryptobiosis.
Cupriavidus metallidurans is a non-spore-forming, Gram-negative bacterium which is adapted to survive several forms of heavy metal stress.
Delftia lacustris is a Gram-negative, nonfermentative, motile, rod-shaped bacterium from the family Comamonadaceae, which was isolated from mesotrophic lake water in Denmark. It has the ability to degrade peptidoglycan through chitinase and lysozyme activity.

Desulfovibrio alaskensis belongs to the sulfate-reducing bacteria. The type strain is Al1T.
Arsenate-reducing bacteria are bacteria which reduce arsenates. Arsenate-reducing bacteria are ubiquitous in arsenic-contaminated groundwater (aqueous environment). Arsenates are salts or esters of arsenic acid (H3AsO4), consisting of the ion AsO43−. They are moderate oxidizers that can be reduced to arsenites and to arsine. Arsenate can serve as a respiratory electron acceptor for oxidation of organic substrates and H2S or H2. Arsenates occur naturally in minerals such as adamite, alarsite, legrandite, and erythrite, and as hydrated or anhydrous arsenates. Arsenates are similar to phosphates since arsenic (As) and phosphorus (P) occur in group 15 (or VA) of the periodic table. Unlike phosphates, arsenates are not readily lost from minerals due to weathering. They are the predominant form of inorganic arsenic in aqueous aerobic environments. On the other hand, arsenite is more common in anaerobic environments, more mobile, and more toxic than arsenate. Arsenite is 25–60 times more toxic and more mobile than arsenate under most environmental conditions. Arsenate can lead to poisoning, since it can replace inorganic phosphate in the glyceraldehyde-3-phosphate --> 1,3-biphosphoglycerate step of glycolysis, producing 1-arseno-3-phosphoglycerate instead. Although glycolysis continues, 1 ATP molecule is lost. Thus, arsenate is toxic due to its ability to uncouple glycolysis. Arsenate can also inhibit pyruvate conversion into acetyl-CoA, thereby blocking the TCA cycle, resulting in additional loss of ATP.
Desulfovibrio oxyclinae is a bacterium. It is sulfate-reducing, and was first isolated from the upper 3mm layer of a hypersaline cyanobacterial mat in Sinai.

The Center for Biofilm Engineering (CBE) is an interdisciplinary research, education, and technology transfer institution located on the central campus of Montana State University in Bozeman, Montana. The center was founded in April 1990 as the Center for Interfacial Microbial Process Engineering with a grant from the Engineering Research Centers (ERC) program of the National Science Foundation (NSF). The CBE integrates faculty from multiple university departments to lead multidisciplinary research teams—including graduate and undergraduate students—to advance fundamental biofilm knowledge, develop beneficial uses for microbial biofilms, and find solutions to industrially relevant biofilm problems. The center tackles biofilm issues including chronic wounds, bioremediation, and microbial corrosion through cross-disciplinary research and education among engineers, microbiologists and industry.
Desulfovibrio desulfuricans is a Gram-negative sulfate-reducing bacteria. It is generally found in soil, water, and the stools of animals, although in rare cases it has been found to cause infection in humans. It is particularly noted for its ability to produce methyl mercury. The reductive glycine pathway, a seventh route for organisms to capture CO2, was discovered in this species. Since these bacteria are killed by exposure to atmospheric oxygen, the environmental niches most frequently occupied by these bacteria are anaerobic. Desulfovibrio desulfuricans 27774 was reported to produce gene transfer agents.
Mercury methylation is the process of forming methylmercury (MeHg). The methylation of mercury can occur abiotically or biotically. Biotically, the primary methylators of mercury are sulfate-reducing and iron-reducing bacteria. Three mechanisms have been proposed for the biotic methylation of mercury by sulfate-reducing bacteria. Mercury methylation can be problematic as methylmercury is toxic and can be bio-magnified through the food web.












