
Yersinia pestis is a gram-negative, non-motile, coccobacillus bacterium without spores that is related to both Yersinia enterocolitica and Yersinia pseudotuberculosis, the pathogen from which Y. pestis evolved and responsible for the Far East scarlet-like fever. It is a facultative anaerobic organism that can infect humans via the Oriental rat flea. It causes the disease plague, which caused the Plague of Justinian and the Black Death, the deadliest pandemic in recorded history. Plague takes three main forms: pneumonic, septicemic, and bubonic. Yersinia pestis is a parasite of its host, the rat flea, which is also a parasite of rats, hence Y. pestis is a hyperparasite.
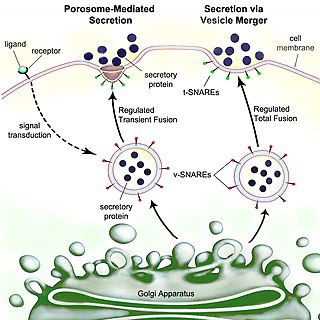
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.

Yersinia pseudotuberculosis is a Gram-negative bacterium that causes Far East scarlet-like fever in humans, who occasionally get infected zoonotically, most often through the food-borne route. Animals are also infected by Y. pseudotuberculosis. The bacterium is urease positive.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:
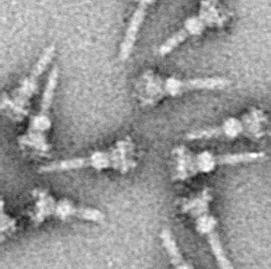
The type III secretion system, also called the injectisome, is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.
Autoinducers are signaling molecules that are produced in response to changes in cell-population density. As the density of quorum sensing bacterial cells increases so does the concentration of the autoinducer. Detection of signal molecules by bacteria acts as stimulation which leads to altered gene expression once the minimal threshold is reached. Quorum sensing is a phenomenon that allows both Gram-negative and Gram-positive bacteria to sense one another and to regulate a wide variety of physiological activities. Such activities include symbiosis, virulence, motility, antibiotic production, and biofilm formation. Autoinducers come in a number of different forms depending on the species, but the effect that they have is similar in many cases. Autoinducers allow bacteria to communicate both within and between different species. This communication alters gene expression and allows bacteria to mount coordinated responses to their environments, in a manner that is comparable to behavior and signaling in higher organisms. Not surprisingly, it has been suggested that quorum sensing may have been an important evolutionary milestone that ultimately gave rise to multicellular life forms.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

Aerobactin is a bacterial iron chelating agent (siderophore) found in E. coli. It is a virulence factor enabling E. coli to sequester iron in iron-poor environments such as the urinary tract.

In molecular biology, the haemolysin expression modulating protein family is a family of proteins. This family consists of haemolysin expression modulating protein (Hha) from Escherichia coli and its enterobacterial homologues, such as YmoA from Yersinia enterocolitica, and RmoA encoded on the R100 plasmid. These proteins act as modulators of bacterial gene expression. Members of this family act in conjunction with members of the H-NS family, participating in the thermoregulation of different virulence factors and in plasmid transfer. Hha, along with the chromatin-associated protein H-NS, is involved in the regulation of expression of the toxin alpha-haemolysin in response to osmolarity and temperature. YmoA modulates the expression of various virulence factors, such as Yop proteins and YadA adhesin, in response to temperature. RmoA is a plasmid R100 modulator involved in plasmid transfer. The HHA family of proteins display striking similarity to the oligomerisation domain of the H-NS proteins.

Rhamnolipids are a class of glycolipid produced by Pseudomonas aeruginosa, amongst other organisms, frequently cited as bacterial surfactants. They have a glycosyl head group, in this case a rhamnose moiety, and a 3-(hydroxyalkanoyloxy)alkanoic acid (HAA) fatty acid tail, such as 3-hydroxydecanoic acid.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).

In molecular biology, the protein domain YopE refers to the secretion of virulence factors in Gram-negative bacteria involves transportation of the protein across two membranes to reach the cell exterior. It not only infects the host cell but also protects the bacteria. It undergoes several mechanisms to evade the host's immune system. This particular protein domain can be referred to as a Rho GTPase-activating protein (GAP).

In molecular biology, YopR is a protein domain commonly found in gram negative bacteria, in particular Yersinia and is a core domain. Proteins in this entry are type III secretion system effectors. They are named differently in different species and in Yersinia has been designated YopR which is encoded by the YscH gene. This Yop protein is unusual in that it is released to the extracellular environment rather than injected directly into the target cell as are most Yop proteins. A hallmark of Yersinia type III machines is the presence of needles extending from the bacterial surface. Needles perform two functions, firstly, as a channel to export effectors into the immune cells and secondly as a sensor.

In molecular biology, the protein domain TyeA is short for Translocation of Yops into eukaryotic cells A. It controls the release of Yersinia outer proteins (Yops) which help Yersinia evade the immune system. More specifically, it interacts with the bacterial protein YopN via hydrophobic residues located on the helices.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.
Bacterial effectors are proteins secreted by pathogenic bacteria into the cells of their host, usually using a type 3 secretion system (TTSS/T3SS), a type 4 secretion system (TFSS/T4SS) or a Type VI secretion system (T6SS). Some bacteria inject only a few effectors into their host’s cells while others may inject dozens or even hundreds. Effector proteins may have many different activities, but usually help the pathogen to invade host tissue, suppress its immune system, or otherwise help the pathogen to survive. Effector proteins are usually critical for virulence. For instance, in the causative agent of plague, the loss of the T3SS is sufficient to render the bacteria completely avirulent, even when they are directly introduced into the bloodstream. Gram negative microbes are also suspected to deploy bacterial outer membrane vesicles to translocate effector proteins and virulence factors via a membrane vesicle trafficking secretory pathway, in order to modify their environment or attack/invade target cells, for example, at the host-pathogen interface.
Daniel A. Portnoy is a microbiologist, the Edward E. Penhoet Distinguished Chair in Global Public Health and Infectious Diseases, and a Professor of Biochemistry, Biophysics and Structural Biology in the Department of Molecular and Cell Biology and in the Division of Microbiology in the Department of Plant and Microbial Biology at the University of California, Berkeley. He is one of the world's foremost experts on Listeria monocytogenes, the bacterium that causes the severe foodborne illness Listeriosis. He has made seminal contributions to multiple aspects of bacterial pathogenesis, cell biology, innate immunity, and cell mediated immunity using L. monocytogenes as a model system and has helped to push forward the use of attenuated L. monocytogenes as an immunotherapeutic tool in the treatment of cancer.

RNA thermometers regulate gene expression in response to temperature allowing pathogens like Yersinia to switch on silent genes after entering the host organism. Usually, RNA thermometers are located in the 5'UTR, but an intergenic RNA thermometer was found in Yersinia pseudotuberculosis. The LcrFRNA thermometer together with the thermo-labile YmoA protein activates synthesis of the most crucial virulence activator LcrF (VirF). The RNA thermosensor sequence is 100% identical in all human pathogenic Yersinia species.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.

Virginia L. Miller is a microbiologist known for her work on studying the factors leading to disease caused by bacteria. Miller is an elected fellow of the American Academy of Microbiology (2003) and a former Pew Charitable Trust Biomedical Scholar (1989).
















