
Escherichia coli ( ESH-ə-RIK-ee-ə KOH-ly) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Treponema pallidum, formerly known as Spirochaeta pallida, is a microaerophilic spirochaete bacterium with subspecies that cause the diseases syphilis, bejel, and yaws. It is transmitted only among humans. It is a helically coiled microorganism usually 6–15 μm long and 0.1–0.2 μm wide. T. pallidum's lack of either a tricarboxylic acid cycle or oxidative phosphorylation results in minimal metabolic activity. The treponemes have a cytoplasmic and an outer membrane. Using light microscopy, treponemes are visible only by using dark-field illumination. T. pallidum consists of three subspecies, T. p. pallidum, T. p. endemicum, and T. p. pertenue, each of which has a distinct associated disease.

Lactococcus lactis is a gram-positive bacterium used extensively in the production of buttermilk and cheese, but has also become famous as the first genetically modified organism to be used alive for the treatment of human disease. L. lactis cells are cocci that group in pairs and short chains, and, depending on growth conditions, appear ovoid with a typical length of 0.5 - 1.5 µm. L. lactis does not produce spores (nonsporulating) and are not motile (nonmotile). They have a homofermentative metabolism, meaning they produce lactic acid from sugars. They've also been reported to produce exclusive L-(+)-lactic acid. However, reported D-(−)-lactic acid can be produced when cultured at low pH. The capability to produce lactic acid is one of the reasons why L. lactis is one of the most important microorganisms in the dairy industry. Based on its history in food fermentation, L. lactis has generally recognized as safe (GRAS) status, with few case reports of it being an opportunistic pathogen.

Leptospirosis is a blood infection caused by the bacteria Leptospira that can infect humans, dogs, rodents and many other wild and domesticated animals. Signs and symptoms can range from none to mild to severe. Weil's disease, the acute, severe form of leptospirosis, causes the infected individual to become jaundiced, develop kidney failure, and bleed. Bleeding from the lungs associated with leptospirosis is known as severe pulmonary haemorrhage syndrome.

Legionella pneumophila is an aerobic, pleomorphic, flagellated, non-spore-forming, Gram-negative bacterium of the genus Legionella. L. pneumophila is the primary human pathogenic bacterium in this group. In nature, L. pneumophila infects freshwater and soil amoebae of the genera Acanthamoeba and Naegleria. This pathogen is found commonly near freshwater environments and will then invade the amoebae found in these environments, using them to carry out metabolic functions.

Nitrosomonas is a genus of Gram-negative bacteria, belonging to the Betaproteobacteria. It is one of the five genera of ammonia-oxidizing bacteria and, as an obligate chemolithoautotroph, uses ammonia as an energy source and carbon dioxide as a carbon source in presence of oxygen. Nitrosomonas are important in the global biogeochemical nitrogen cycle, since they increase the bioavailability of nitrogen to plants and in the denitrification, which is important for the release of nitrous oxide, a powerful greenhouse gas. This microbe is photophobic, and usually generate a biofilm matrix, or form clumps with other microbes, to avoid light. Nitrosomonas can be divided into six lineages: the first one includes the species Nitrosomonas europea, Nitrosomonas eutropha, Nitrosomonas halophila, and Nitrosomonas mobilis. The second lineage presents the species Nitrosomonas communis, N. sp. I and N. sp. II, meanwhile the third lineage includes only Nitrosomonas nitrosa. The fourth lineage includes the species Nitrosomonas ureae and Nitrosomonas oligotropha and the fifth and sixth lineages include the species Nitrosomonas marina, N. sp. III, Nitrosomonas estuarii and Nitrosomonas cryotolerans.

Leptospira is a genus of spirochaete bacteria, including a small number of pathogenic and saprophytic species. Leptospira was first observed in 1907 in kidney tissue slices of a leptospirosis victim who was described as having died of "yellow fever".
Leptospira noguchii is a gram-negative, pathogenic organism named for Japanese bacteriologist Dr. Hideyo Noguchi who named the genus Leptospira. L. noguchii is famous for causing the febrile illness in Fort Bragg, NC during World War II. There was 40 cases of this fever documented during each summer from 1942 to 1944; however, there were 0 deaths recorded from this outbreak. Unlike other strains of Leptospira that cause leptospirosis, L. noguchii is characterized by showing a pretibial rash on the victim. Its specific epithet recognises Hideyo Noguchi.

Leptospira interrogans is a species of obligate aerobic spirochaete bacteria shaped like a corkscrew with hooked and spiral ends. L. interrogans is mainly found in warmer tropical regions. The bacteria can live for weeks to months in the ground or water. Leptospira is one of the genera of the spirochaete phylum that causes severe mammalian infections. This species is pathogenic to some wild and domestic animals, including pet dogs. It can also spread to humans through abrasions on the skin, where infection can cause flu-like symptoms with kidney and liver damage. Human infections are commonly spread by contact with contaminated water or soil, often through the urine of both wild and domestic animals. Some individuals are more susceptible to serious infection, including farmers and veterinarians who work with animals.
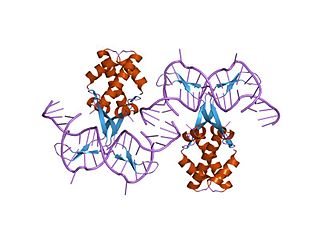
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria and archaea, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.
Leptospira kirschneri is a Gram negative, obligate aerobe species of spirochete bacteria named for University of Otago bacteriologist Dr. Leopold Kirschner. It is a member of the genus Leptospira. The species is pathogenic and can cause leptospirosis, most commonly in pigs.
Leptospira broomii is a species of Leptospira isolated from humans with leptospirosis. The type strain is 5399T.
Spiral bacteria, bacteria of spiral (helical) shape, form the third major morphological category of prokaryotes along with the rod-shaped bacilli and round cocci. Spiral bacteria can be subclassified by the number of twists per cell, cell thickness, cell flexibility, and motility. The two types of spiral cells are spirillum and spirochete, with spirillum being rigid with external flagella, and spirochetes being flexible with internal flagella.
The gene rpoN encodes the sigma factor sigma-54, a protein in Escherichia coli and other species of bacteria. RpoN antagonizes RpoS sigma factors.

Bacteroides thetaiotaomicron is a gram-negative, rod shaped obligate anaerobic bacterium that is a prominent member of the normal gut microbiome in the distal intestines. Its proteome, consisting of 4,779 members, includes a system for obtaining and breaking down dietary polysaccharides that would otherwise be difficult to digest. B. thetaiotaomicron is also an opportunistic pathogen, meaning it may become virulent in immunocompromised individuals. It is often used in research as a model organism for functional studies of the human microbiota.
Leptospira alstonii is a gram negative, mobile, spirochete. It is flexible, helical, and motile by means of two periplasmic flagella. It is obligately aerobic and oxidase positive. It was named after J. M. Alston, a British microbiologist who made significant contributions to the study of Leptospirosis. It is one of nine human or animal pathogenic species of Leptospira. It was originally isolated from material submitted to the Veterinary Diagnostic Laboratory at Iowa State University during an outbreak of swine abortion in 1983. It has been isolated and stored in liquid nitrogen or Ellinghausen-McCullough-Johnson-Harris medium. It also has been isolated in China from a frog. The strain is also available from culture collections of the WHO collaborating centers. Lipase is not produced by this species. NaCl is not required for growth. Growth is inhibited by 8-azaguanine at 225 µg/mL or 2,6-diaminopurine (10 µg/mL) and copper sulfate. It contains serovars from the serogroup ranarum. DNA G+C content is 39±8 mol%.
Leopold Kirschner was an Austro-Hungarian, Dutch, and New Zealand bacteriologist specializing in leptospirosis. He is known for his work on the survival of Leptospira spp in the environment, research on conditions and media for Leptospira growth, his role in the initial discoveries of leptospirosis in New Zealand, for early epidemiologic descriptions of leptospirosis as an occupational disease of dairy farmers, and for the major pathogenic Leptospira species, Leptospira kirschneri, that was named in his honor.
Joseph Michael Vinetz is a Professor of Medicine and Anthropology at Yale University, Research Professor at the Universidad Peruana Cayetano Heredia and Associate Investigator of the Alexander von Humboldt Institute of Tropical Medicine at the Universidad Peruana Cayetano Heredia.
Leptospira wolffii is a gram negative aerobic bacterium in the spirochaete phylum. The species named after Dutch bacteriologist Jan Willem Wolff.
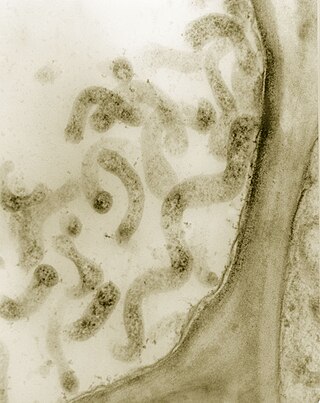
Spiroplasma kunkelii is a species of Mollicutes, which are small bacteria that all share a common cell wall-less feature. They are characterized by helical and spherical morphology, they actually have the ability to be spherical or helical depending on the circumstances. The cells movement is bound by a membrane. The cell size ranges from 0.15 to 0.20 micrometers.









