
Leptospirosis is a blood infection caused by the bacteria Leptospira that can infect humans, dogs, rodents and many other wild and domesticated animals. Signs and symptoms can range from none to mild to severe. Weil's disease, the acute, severe form of leptospirosis, causes the infected individual to become jaundiced, develop kidney failure, and bleed. Bleeding from the lungs associated with leptospirosis is known as severe pulmonary haemorrhage syndrome.

Yersinia pseudotuberculosis is a Gram-negative bacterium that causes Far East scarlet-like fever in humans, who occasionally get infected zoonotically, most often through the food-borne route. Animals are also infected by Y. pseudotuberculosis. The bacterium is urease positive.
Chlamydia muridarum is an intracellular bacterial species that at one time belonged to Chlamydia trachomatis. However, C. trachomatis naturally only infects humans and C. muridarum naturally infects only members of the family Muridae.

N-Acetylneuraminic acid is the predominant sialic acid found in human cells, and many mammalian cells. Other forms, such as N-Glycolylneuraminic acid, may also occur in cells.

Sporothrix schenckii, a fungus that can be found worldwide in the environment, is named for medical student Benjamin Schenck, who in 1896 was the first to isolate it from a human specimen. The species is present in soil as well as in and on living and decomposing plant material such as peat moss. It can infect humans as well as animals and is the causative agent of sporotrichosis, commonly known as "rose handler's disease." The most common route of infection is the introduction of spores to the body through a cut or puncture wound in the skin. Infection commonly occurs in otherwise healthy individuals but is rarely life-threatening and can be treated with antifungals. In the environment it is found growing as filamentous hyphae. In host tissue it is found as a yeast. The transition between the hyphal and yeast forms is temperature dependent making S. schenckii a thermally dimorphic fungus.
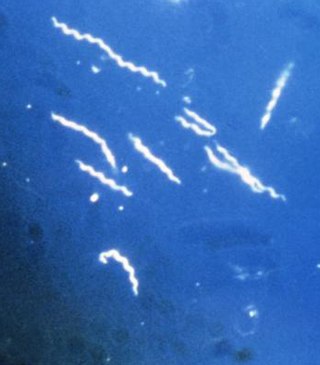
Lyme disease, or borreliosis, is caused by spirochetal bacteria from the genus Borrelia, which has 52 known species. Three main species are the main causative agents of the disease in humans, while a number of others have been implicated as possibly pathogenic. Borrelia species in the species complex known to cause Lyme disease are collectively called Borrelia burgdorferisensu lato (s.l.) not to be confused with the single species in that complex Borrelia burgdorferi sensu stricto which is responsible for nearly all cases of Lyme disease in North America.
Listeriolysin O (LLO) is a hemolysin produced by the bacterium Listeria monocytogenes, the pathogen responsible for causing listeriosis. The toxin may be considered a virulence factor, since it is crucial for the virulence of L. monocytogenes.
Leptospira noguchii is a gram-negative, pathogenic organism named for Japanese bacteriologist Dr. Hideyo Noguchi who named the genus Leptospira. L. noguchii is famous for causing the febrile illness in Fort Bragg, NC during World War II. There was 40 cases of this fever documented during each summer from 1942 to 1944; however, there were 0 deaths recorded from this outbreak. Unlike other strains of Leptospira that cause leptospirosis, L. noguchii is characterized by showing a pretibial rash on the victim. Its specific epithet recognises Hideyo Noguchi.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

Leptospira interrogans is a species of obligate aerobic spirochaete bacteria shaped like a corkscrew with hooked and spiral ends. L. interrogans is mainly found in warmer tropical regions. The bacteria can live for weeks to months in the ground or water. Leptospira is one of the genera of the spirochaete phylum that causes severe mammalian infections. This species is pathogenic to some wild and domestic animals, including pet dogs. It can also spread to humans through abrasions on the skin, where infection can cause flu-like symptoms with kidney and liver damage. Human infections are commonly spread by contact with contaminated water or soil, often through the urine of both wild and domestic animals. Some individuals are more susceptible to serious infection, including farmers and veterinarians who work with animals.

The AB toxins are two-component protein complexes secreted by a number of pathogenic bacteria, though there is a pore-forming AB toxin found in the eggs of a snail. They can be classified as Type III toxins because they interfere with internal cell function. They are named AB toxins due to their components: the "A" component is usually the "active" portion, and the "B" component is usually the "binding" portion. The "A" subunit possesses enzyme activity, and is transferred to the host cell following a conformational change in the membrane-bound transport "B" subunit. These proteins consist of two independent polypeptides, which correspond to the A/B subunit moieties. The enzyme component (A) enters the cell through endosomes produced by the oligomeric binding/translocation protein (B), and prevents actin polymerisation through ADP-ribosylation of monomeric G-actin.

VrrA is a non-coding RNA that is conserved across all Vibrio species of bacteria and acts as a repressor for the synthesis of the outer membrane protein OmpA. This non-coding RNA was initially identified from Tn5 transposon mutant libraries of Vibrio cholerae and its location within the bacterial genome was mapped to the intergenic region between genes VC1741 and VC1743 by RACE analysis.
Paracytophagy is the cellular process whereby a cell engulfs a protrusion which extends from a neighboring cell. This protrusion may contain material which is actively transferred between the cells. The process of paracytophagy was first described as a crucial step during cell-to-cell spread of the intracellular bacterial pathogen Listeria monocytogenes, and is also commonly observed in Shigella flexneri. Paracytophagy allows these intracellular pathogens to spread directly from cell to cell, thus escaping immune detection and destruction. Studies of this process have contributed significantly to our understanding of the role of the actin cytoskeleton in eukaryotic cells.
Leptospira kirschneri is a Gram negative, obligate aerobe species of spirochete bacteria named for University of Otago bacteriologist Dr. Leopold Kirschner. It is a member of the genus Leptospira. The species is pathogenic and can cause leptospirosis, most commonly in pigs.
Leptospira broomii is a species of Leptospira isolated from humans with leptospirosis. The type strain is 5399T.
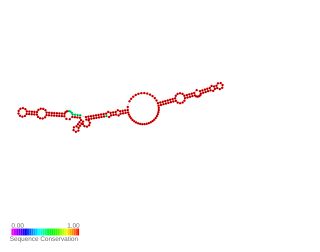
Lig RNA thermometer is a cis-acting non-coding RNA element that controls ligA and ligB gene expression in Leptospira interrogans in response to temperature change. The lipoproteins LigA and LigB stimulate adhesion of the element and then hosting proteins. The RNA that composes of 175-nucleotide 5'UTR and the first six lig codons folds into two distinct-stem loop structures. Lig expression is limited by these double-stranded RNA structures because they occludes the ribosome-binding site. At higher temperatures, the ribosome binding site is exposed to promote translation initiation.
Leptospira alstonii is a gram negative, mobile, spirochete. It is flexible, helical, and motile by means of two periplasmic flagella. It is obligately aerobic and oxidase positive. It was named after J. M. Alston, a British microbiologist who made significant contributions to the study of Leptospirosis. It is one of nine human or animal pathogenic species of Leptospira. It was originally isolated from material submitted to the Veterinary Diagnostic Laboratory at Iowa State University during an outbreak of swine abortion in 1983. It has been isolated and stored in liquid nitrogen or Ellinghausen-McCullough-Johnson-Harris medium. It also has been isolated in China from a frog. The strain is also available from culture collections of the WHO collaborating centers. Lipase is not produced by this species. NaCl is not required for growth. Growth is inhibited by 8-azaguanine at 225 µg/mL or 2,6-diaminopurine (10 µg/mL) and copper sulfate. It contains serovars from the serogroup ranarum. DNA G+C content is 39±8 mol%.
Joseph Michael Vinetz is a Professor of Medicine and Anthropology at Yale University, Research Professor at the Universidad Peruana Cayetano Heredia and Associate Investigator of the Alexander von Humboldt Institute of Tropical Medicine at the Universidad Peruana Cayetano Heredia.
Leptospira biflexa is a spirochaete bacterium in the genus Leptospira and was the first saprophytic Leptospira genome to be sequenced.










