Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

Prolyl endopeptidase (PE) also known as prolyl oligopeptidase or post-proline cleaving enzyme is an enzyme that in humans is encoded by the PREP gene.

Ficain also known as ficin, debricin, or higueroxyl delabarre is a proteolytic enzyme extracted from the latex sap from the stems, leaves, and unripe fruit of the American wild fig tree Ficus insipida.
Leumorphin, also known as dynorphin B1–29, is a naturally occurring endogenous opioid peptide. Derived as a proteolytic cleavage product of residues 226-254 of prodynorphin, leumorphin is a nonacosapeptide and has the sequence Tyr-Gly-Gly-Phe-Leu-Arg-Arg-Gln-Phe-Lys-Val-Val-Thr-Arg-Ser-Gln-Glu-Asp-Pro-Asn-Ala-Tyr-Ser-Gly-Glu-Leu-Phe-Asp-Ala. It can be further reduced to dynorphin B and dynorphin B-14 by pitrilysin metallopeptidase 1, an enzyme of the endopeptidase family. Leumorphin behaves as a potent and selective κ-opioid receptor agonist, similarly to other endogenous opioid peptide derivatives of prodynorphin.

Glutamyl endopeptidase is an extracellular bacterial serine protease of the glutamyl endopeptidase I family that was initially isolated from the Staphylococcus aureus strain V8. The protease is, hence, commonly referred to as "V8 protease", or alternatively SspA from its corresponding gene.
Lysyl endopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
C-terminal processing peptidase is an enzyme. This enzyme catalyses the following chemical reaction
Adenain is an enzyme. This enzyme catalyses the following chemical reaction
Nuclear-inclusion-a endopeptidase is a protease enzyme found in potyviruses. This enzyme catalyses the following chemical reaction:
Helper-component proteinase is an enzyme. This enzyme catalyses the following chemical reaction
Staphopain is an enzyme. This enzyme catalyses the following chemical reaction
Calicivirin is an enzyme. This enzyme catalyses the following chemical reaction
Ulp1 peptidase is an enzyme. This enzyme catalyses the following chemical reaction

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.
Asparagine peptide lyase are one of the seven groups in which proteases, also termed proteolytic enzymes, peptidases, or proteinases, are classified according to their catalytic residue. The catalytic mechanism of the asparagine peptide lyases involves an asparagine residue acting as nucleophile to perform a nucleophilic elimination reaction, rather than hydrolysis, to catalyse the breaking of a peptide bond.
Glutamyl endopeptidase I is a family of extracellular bacterial serine proteases. The proteases within this family have been identified in species of Staphylococcus, Bacillus, and Streptomyces, among others. The two former are more closely related, while the Streptomyces-type is treated as a separate family, glutamyl endopeptidase II.
Flock House virus (FHV) is in the alphanodavirus genus of the Nodaviridae family of viruses. Flock House virus was isolated from a grass grub at the Flock House research station in Bulls, New Zealand. FHV is an extensively studied virus and is considered a model system for the study of other non-enveloped RNA viruses owing to its small size and genetic tractability, particularly to study the role of the transiently exposed hydrophobic gamma peptide and the metastability of the viral capsid. FHV can be engineered in insect cell culture allowing for the tailored production of native or mutant authentic virions or virus-like-particles. FHV is a platform for nanotechnology and nanomedicine, for example, for epitope display and vaccine development. Viral entry into host cells occurs via receptor-mediated endocytosis. Receptor binding initiates a sequence of events during which the virus exploits the host environment in order to deliver the viral cargo in to the host cytosol. Receptor binding prompts the meta-stability of the capsid–proteins, the coordinated rearrangements of which are crucial for subsequent steps in the infection pathway. In addition, the transient exposure of a covalently-independent hydrophobic γ-peptide is responsible for breaching cellular membranes and is thus essential for the viral entry of FHV into host cells.
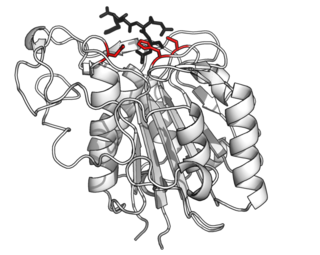
Asparagine endopeptidase is a proteolytic enzyme from C13 peptidase family which hydrolyses a peptide bond using the thiol group of a cysteine residue as a nucleophile. It is also known as asparaginyl endopeptidase, citvac, proteinase B, hemoglobinase, PRSC1 gene product or LGMN, vicilin peptidohydrolase and bean endopeptidase. In humans it is encoded by the LGMN gene.
References
- ↑ Zlotnick A, Reddy VS, Dasgupta R, Schneemann A, Ray WJ, Rueckert RR, Johnson JE (May 1994). "Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue". The Journal of Biological Chemistry. 269 (18): 13680–4. PMID 8175803.
- ↑ Johnson JE, Schneemann A (1998). "Nodavirus endopeptidase". In Barrett AJ, Rawlings ND, Woessner JF (eds.). Handbook of Proteolytic Enzymes. London: Handbook of Proteolytic Enzymes. pp. 964–967.