Related Research Articles

The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside, including the skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, biliary tract, and gastrointestinal tract. Types of human microbiota include bacteria, archaea, fungi, protists, and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term human microbiome is sometimes used to refer to the collective genomes of resident microorganisms; however, the term human metagenome has the same meaning.

The Bacillota are a phylum of bacteria, most of which have gram-positive cell wall structure. The renaming of phyla such as Firmicutes in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature.

Gut microbiota, gut microbiome, or gut flora, are the microorganisms, including bacteria, archaea, fungi, and viruses, that live in the digestive tracts of animals. The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota. The gut is the main location of the human microbiome. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis.

Bacteroides is a genus of Gram-negative, obligate anaerobic bacteria. Bacteroides species are non endospore-forming bacilli, and may be either motile or nonmotile, depending on the species. The DNA base composition is 40–48% GC. Unusual in bacterial organisms, Bacteroides membranes contain sphingolipids. They also contain meso-diaminopimelic acid in their peptidoglycan layer.

Paneth cells are cells in the small intestine epithelium, alongside goblet cells, enterocytes, and enteroendocrine cells. Some can also be found in the cecum and appendix. They are located below the intestinal stem cells in the intestinal glands and the large eosinophilic refractile granules that occupy most of their cytoplasm.

Germ-free organisms are multi-cellular organisms that have no microorganisms living in or on them. Such organisms are raised using various methods to control their exposure to viral, bacterial or parasitic agents. When known microbiota are introduced to a germ-free organism, it usually is referred to as a gnotobiotic organism, however technically speaking, germ-free organisms are also gnotobiotic because the status of their microbial community is known. Due to lacking a microbiome, many germ-free organisms exhibit health deficits such as defects in the immune system and difficulties with energy acquisition. Typically germ-free organisms are used in the study of a microbiome where careful control of outside contaminants is required.
Dysbiosis is characterized by a disruption to the microbiome resulting in an imbalance in the microbiota, changes in their functional composition and metabolic activities, or a shift in their local distribution. For example, a part of the human microbiota such as the skin flora, gut flora, or vaginal flora, can become deranged, with normally dominating species underrepresented and normally outcompeted or contained species increasing to fill the void. Dysbiosis is most commonly reported as a condition in the gastrointestinal tract.
Microecology means microbial ecology or ecology of a microhabitat. In humans, gut microecology is the study of the microbial ecology of the human gut which includes gut microbiota composition, its metabolic activity, and the interactions between the microbiota, the host, and the environment. Research in human gut microecology is important because the microbiome can have profound effects on human health. The microbiome is known to influence the immune system, digestion, and metabolism, and is thought to play a role in a variety of diseases, including diabetes, obesity, inflammatory bowel disease, and cancer. Studying the microbiome can help us better understand these diseases and develop treatments.
Rikenellaceae is a family of Gram-negative bacteria described by Noel R. Krieg in 2015. It contains nine genera, five of which are validly published by the International Code of Nomenclature of Prokaryotes. Bacteria with 16S ribosomal RNA highly similar to the Rikenella genus, as compared to the larger taxonomic order Bacteroidales, are classified in this family.
Jeffrey Ivan Gordon is a biologist and the Dr. Robert J. Glaser Distinguished University Professor and Director of the Center for Genome Sciences and Systems Biology at Washington University in St. Louis. He is internationally known for his research on gastrointestinal development and how gut microbial communities affect normal intestinal function, shape various aspects of human physiology including our nutritional status, and affect predisposition to diseases. He is a member of the National Academy of Sciences, the American Academy of Arts and Sciences, the Institute of Medicine of the National Academies, and the American Philosophical Society.

Long-term close-knit interactions between symbiotic microbes and their host can alter host immune system responses to other microorganisms, including pathogens, and are required to maintain proper homeostasis. The immune system is a host defense system consisting of anatomical physical barriers as well as physiological and cellular responses, which protect the host against harmful microorganisms while limiting host responses to harmless symbionts. Humans are home to 1013 to 1014 bacteria, roughly equivalent to the number of human cells, and while these bacteria can be pathogenic to their host most of them are mutually beneficial to both the host and bacteria.

Microbiota are the range of microorganisms that may be commensal, mutualistic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses, and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.
Faecalibacterium is a genus of bacteria. The genus contains several species including Faecalibacterium prausnitzii, Faecalibacterium butyricigenerans, Faecalibacterium longum, Faecalibacterium duncaniae, Faecalibacterium hattorii, and Faecalibacterium gallinarum. Its first known species, Faecalibacterium prausnitzii is gram-positive, mesophilic, rod-shaped, and anaerobic, and is one of the most abundant and important commensal bacteria of the human gut microbiota. It is non-spore forming and non-motile. These bacteria produce butyrate and other short-chain fatty acids through the fermentation of dietary fiber. The production of butyrate makes them an important member of the gut microbiota, fighting against inflammation.
Ruminococcus is a genus of bacteria in the class Clostridia. They are anaerobic, Gram-positive gut microbes. One or more species in this genus are found in significant numbers in the human gut microbiota. The type species is R. flavefaciens. As usual, bacteria taxonomy is in flux, with Clostridia being paraphyletic, and some erroneous members of Ruminococcus being reassigned to a new genus Blautia on the basis of 16S rRNA gene sequences.
Sutterella is a genus of Gram-negative, rod-shaped, non-spore-forming, Betaproteobacteria whose species have been isolated from the human gastrointestinal tract as well as canine feces. The genus of the family Sutterellaceae currently encompasses 4 distinct species, though at least 5 additional species have been proposed that do not yet meet International Code of Nomenclature of Prokaryotes (ICNP) standards for classification. Sutterella are frequently referred to as commensal in the context of human hosts, but are associated with inflammation, which has implications for a number of diseases.

Akkermansia muciniphila is a human intestinal symbiont, isolated from human feces. It is a mucin-degrading bacterium belonging to the genus, Akkermansia, discovered in 2004 by Muriel Derrien and Willem de Vos at Wageningen University of the Netherlands. It belongs to the phylum Verrucomicrobiota and its type strain is MucT. It is under preliminary research for its potential association with metabolic disorders.

The gut–brain axis is the two-way biochemical signaling that takes place between the gastrointestinal tract and the central nervous system (CNS). The "microbiota–gut–brain axis" includes the role of gut microbiota in the biochemical signaling events that take place between the GI tract and the CNS. Broadly defined, the gut–brain axis includes the central nervous system, neuroendocrine system, neuroimmune systems, the hypothalamic–pituitary–adrenal axis, sympathetic and parasympathetic arms of the autonomic nervous system, the enteric nervous system, vagus nerve, and the gut microbiota.
Bacteriotherapy is the purposeful use of bacteria or their products in treating an illness. Forms of bacteriotherapy include the use of probiotics, microorganisms that provide health benefits when consumed; fecal matter transplants (FMT) /intestinal microbiota transplant (IMT), the transfer of gut microorganisms from the fecal matter of healthy donors to recipient patients to restore microbiota; or synbiotics which combine prebiotics, indigestible ingredients that promote growth of beneficial microorganisms, and probiotics. Through these methods, the gut microbiota, the community of 300-500 microorganism species that live in the digestive tract of animals aiding in digestion, energy storage, immune function and protection against pathogens, can be recolonized with favorable bacteria, which in turn has therapeutic effects.
The Human Microbiome Project (HMP), completed in 2012, laid the foundation for further investigation into the role the microbiome plays in overall health and disease. One area of particular interest is the role which delivery mode plays in the development of the infant/neonate microbiome and what potential implications this may have long term. It has been found that infants born via vaginal delivery have microbiomes closely mirroring that of the mother's vaginal microbiome, whereas those born via cesarean section tend to resemble that of the mother's skin. One notable study from 2010 illustrated an abundance of Lactobacillus and other typical vaginal genera in stool samples of infants born via vaginal delivery and an abundance of Staphylococcus and Corynebacterium, commonly found on the skin surfaces, in stool samples of infants born via cesarean section. From these discoveries came the concept of vaginal seeding, also known as microbirthing, which is a procedure whereby vaginal fluids are applied to a new-born child delivered by caesarean section. The idea of vaginal seeding was explored in 2015 after Maria Gloria Dominguez-Bello discovered that birth by caesarean section significantly altered the newborn child's microbiome compared to that of natural birth. The purpose of the technique is to recreate the natural transfer of bacteria that the baby gets during a vaginal birth. It involves placing swabs in the mother's vagina, and then wiping them onto the baby's face, mouth, eyes and skin. Due to the long-drawn nature of studying the impact of vaginal seeding, there are a limited number of studies available that support or refute its use. The evidence suggests that applying microbes from the mother's vaginal canal to the baby after cesarean section may aid in the partial restoration of the infant's natural gut microbiome with an increased likelihood of pathogenic infection to the child via vertical transmission.
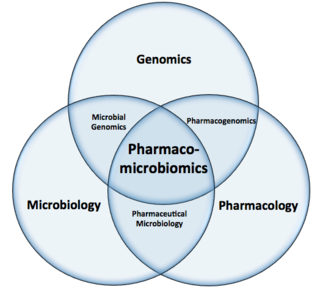
Pharmacomicrobiomics, proposed by Prof. Marco Candela for the ERC-2009-StG project call, and publicly coined for the first time in 2010 by Rizkallah et al., is defined as the effect of microbiome variations on drug disposition, action, and toxicity. Pharmacomicrobiomics is concerned with the interaction between xenobiotics, or foreign compounds, and the gut microbiome. It is estimated that over 100 trillion prokaryotes representing more than 1000 species reside in the gut. Within the gut, microbes help modulate developmental, immunological and nutrition host functions. The aggregate genome of microbes extends the metabolic capabilities of humans, allowing them to capture nutrients from diverse sources. Namely, through the secretion of enzymes that assist in the metabolism of chemicals foreign to the body, modification of liver and intestinal enzymes, and modulation of the expression of human metabolic genes, microbes can significantly impact the ingestion of xenobiotics.
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Nagai, Fumiko; Morotomi, Masami; Sakon, Hiroshi; Tanaka, Ryuichiro (2009). "Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces". International Journal of Systematic and Evolutionary Microbiology. 59 (7): 1793–1797. doi: 10.1099/ijs.0.002519-0 . PMID 19542131.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Morotomi, Masami; Nagai, Fumiko; Watanabe, Yohei (2011). "Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales". International Journal of Systematic and Evolutionary Microbiology. 61 (3): 637–643. doi: 10.1099/ijs.0.023556-0 . PMID 20400667.
- ↑ Jeong, Ju-Yong; Park, Hee-Deung; Lee, Kyong-Hee; Weon, Hang-Yeon; Ka, Jong-Ok (2011-08-01). "Microbial community analysis and identification of alternative host-specific fecal indicators in fecal and river water samples using pyrosequencing". The Journal of Microbiology. 49 (4): 585–94. doi:10.1007/s12275-011-0530-6. ISSN 1225-8873. PMID 21887641. S2CID 43413479.
- 1 2 3 "Search". www.arb-silva.de. Retrieved 2017-11-30.
- ↑ Zhang, Chenhong; Zhang, Menghui; Pang, Xiaoyan; Zhao, Yufeng; Wang, Linghua; Zhao, Liping (October 2012). "Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations". The ISME Journal. 6 (10): 1848–1857. doi:10.1038/ismej.2012.27. ISSN 1751-7370. PMC 3446802 . PMID 22495068.
- ↑ Noble, Emily E.; Hsu, Ted M.; Jones, Roshonda B.; Fodor, Anthony A.; Goran, Michael I.; Kanoski, Scott E. (2017-01-01). "Early-Life Sugar Consumption Affects the Rat Microbiome Independently of Obesity". The Journal of Nutrition. 147 (1): 20–28. doi:10.3945/jn.116.238816. ISSN 0022-3166. PMC 5177734 . PMID 27903830.
- ↑ Zhang, Xiaoxia; Wang, Hao; Yin, Peipei; Fan, Hang; Sun, Liwei; Liu, Yujun (2017-02-22). "Flaxseed oil ameliorates alcoholic liver disease via anti-inflammation and modulating gut microbiota in mice". Lipids in Health and Disease. 16 (1): 44. doi: 10.1186/s12944-017-0431-8 . ISSN 1476-511X. PMC 5322643 . PMID 28228158.
- 1 2 Chiodini, Rodrick J.; Dowd, Scot E.; Chamberlin, William M.; Galandiuk, Susan; Davis, Brian; Glassing, Angela (2015-07-29). "Microbial Population Differentials between Mucosal and Submucosal Intestinal Tissues in Advanced Crohn's Disease of the Ileum". PLOS ONE. 10 (7): e0134382. Bibcode:2015PLoSO..1034382C. doi: 10.1371/journal.pone.0134382 . ISSN 1932-6203. PMC 4519195 . PMID 26222621.
- 1 2 Huang, Chunlan; Chen, Jing; Wang, Jingjing; Zhou, Hui; Lu, Yingying; Lou, Lihong; Zheng, Junyuan; Tian, Ling; Wang, Xingpeng (2017). "Dysbiosis of Intestinal Microbiota and Decreased Antimicrobial Peptide Level in Paneth Cells during Hypertriglyceridemia-Related Acute Necrotizing Pancreatitis in Rats". Frontiers in Microbiology. 8: 776. doi: 10.3389/fmicb.2017.00776 . ISSN 1664-302X. PMC 5415626 . PMID 28522995.
- ↑ Carding, Simon; Verbeke, Kristin; Vipond, Daniel T.; Corfe, Bernard M.; Owen, Lauren J. (2015). "Dysbiosis of the gut microbiota in disease". Microbial Ecology in Health & Disease. 26: 26191. doi:10.3402/mehd.v26.26191. PMC 4315779 . PMID 25651997.