
A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-aca-D-alanyl moiety of R-L-aca-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.

Inhibitors of dipeptidyl peptidase 4 are a class of oral hypoglycemics that block the enzyme dipeptidyl peptidase-4 (DPP-4). They can be used to treat diabetes mellitus type 2.
Collagenases are enzymes that break the peptide bonds in collagen. They assist in destroying extracellular structures in the pathogenesis of bacteria such as Clostridium. They are considered a virulence factor, facilitating the spread of gas gangrene. They normally target the connective tissue in muscle cells and other body organs.
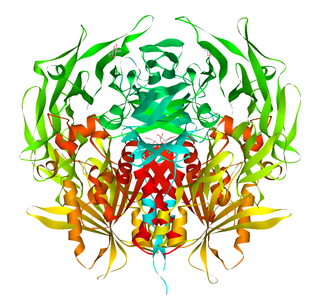
Dipeptidyl peptidase-4 (DPP4), also known as adenosine deaminase complexing protein 2 or CD26 is a protein that, in humans, is encoded by the DPP4 gene. DPP4 is related to FAP, DPP8, and DPP9. The enzyme was discovered in 1966 by Hopsu-Havu and Glenner, and as a result of various studies on chemism, was called dipeptidyl peptidase IV [DP IV].
Transition state analogs, are chemical compounds with a chemical structure that resembles the transition state of a substrate molecule in an enzyme-catalyzed chemical reaction. Enzymes interact with a substrate by means of strain or distortions, moving the substrate towards the transition state. Transition state analogs can be used as inhibitors in enzyme-catalyzed reactions by blocking the active site of the enzyme. Theory suggests that enzyme inhibitors which resembled the transition state structure would bind more tightly to the enzyme than the actual substrate. Examples of drugs that are transition state analog inhibitors include flu medications such as the neuraminidase inhibitor oseltamivir and the HIV protease inhibitors saquinavir in the treatment of AIDS.

Thermolysin is a thermostable neutral metalloproteinase enzyme produced by the Gram-positive bacteria Bacillus thermoproteolyticus. It requires one zinc ion for enzyme activity and four calcium ions for structural stability. Thermolysin specifically catalyzes the hydrolysis of peptide bonds containing hydrophobic amino acids. However thermolysin is also widely used for peptide bond formation through the reverse reaction of hydrolysis. Thermolysin is the most stable member of a family of metalloproteinases produced by various Bacillus species. These enzymes are also termed 'neutral' proteinases or thermolysin -like proteinases (TLPs).
Dipeptidyl peptidase-4 inhibitors are enzyme inhibitors that inhibit the enzyme dipeptidyl peptidase-4 (DPP-4). They are used in the treatment of type 2 diabetes mellitus. Inhibition of the DPP-4 enzyme prolongs and enhances the activity of incretins that play an important role in insulin secretion and blood glucose control regulation. Type 2 diabetes mellitus is a chronic metabolic disease that results from inability of the β-cells in the pancreas to secrete sufficient amounts of insulin to meet the body's needs. Insulin resistance and increased hepatic glucose production can also play a role by increasing the body's demand for insulin. Current treatments, other than insulin supplementation, are sometimes not sufficient to achieve control and may cause undesirable side effects, such as weight gain and hypoglycemia. In recent years, new drugs have been developed, based on continuing research into the mechanism of insulin production and regulation of the metabolism of sugar in the body. The enzyme DPP-4 has been found to play a significant role.

Talopeptin is a chemical compound derived from cultures of Streptomyces. It is a known reversible inhibitor of thermolysin and is expected to inhibit other metalloproteinases. Chemically, talopeptin differs from its closely related peptidase inhibitor phosphoramidon by a single stereocenter.

Ilomastat (INN), is a broad-spectrum matrix metalloproteinase inhibitor.

Ecadotril is a neutral endopeptidase inhibitor and determined by the presence of peptidase family M13 as a neutral endopeptidase inhibited by phosphoramidon. Ecadotril is the (S)-enantiomer of racecadotril. NEP-like enzymes include the endothelin-converting enzymes. The peptidase M13 family believed to activate or inactivate oligopeptide (pro)-hormones such as opioid peptides, neprilysin is another member of this group, in the case of the metallopeptidases and aspartic, the nucleophiles clan or family for example MA, is an activated water molecule. The peptidase domain for members of this family also contains a bacterial member and resembles that of thermolysin the predicted active site residues for members of this family and thermolysin occur in the motif HEXXH. Thermolysin complexed with the inhibitor (S)-thiorphan are isomeric thiol-containing inhibitors of endopeptidase EC 24-11.

Candoxatril is the orally active prodrug of candoxatrilat (UK-73967).
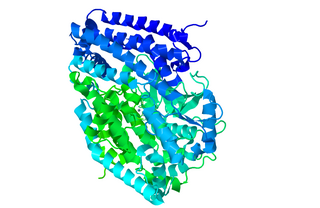
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.

Aspergilloglutamic peptidase, also called aspergillopepsin II is a proteolytic enzyme. The enzyme was previously thought be an aspartic protease, but it was later shown to be a glutamic protease with a catalytic Glu residue at the active site, and was therefore renamed aspergilloglutamic peptidase.

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.
Pseudolysin is an enzyme. This enzyme catalyses the following chemical reaction
An endopeptidase inhibitor is a drug that inhibits one or more endopeptidase enzymes. Endopeptidases are one of two types of proteases, the other being exopeptidases. Endopeptidases cleave peptide bonds of non-terminal amino acids, whereas exopeptidases break terminal bonds, resulting in the release of a single amino acid or dipeptide from the peptide chain.














