
Methionine is an essential amino acid in humans.
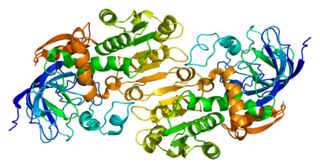
Alcohol dehydrogenases (ADH) (EC 1.1.1.1) are a group of dehydrogenase enzymes that occur in many organisms and facilitate the interconversion between alcohols and aldehydes or ketones with the reduction of nicotinamide adenine dinucleotide (NAD+) to NADH. In humans and many other animals, they serve to break down alcohols that are otherwise toxic, and they also participate in the generation of useful aldehyde, ketone, or alcohol groups during the biosynthesis of various metabolites. In yeast, plants, and many bacteria, some alcohol dehydrogenases catalyze the opposite reaction as part of fermentation to ensure a constant supply of NAD+.

Sulfur assimilation is the process by which living organisms incorporate sulfur into their biological molecules. In plants, sulfate is absorbed by the roots and then transported to the chloroplasts by the transipration stream where the sulfur are reduced to sulfide with the help of a series of enzymatic reactions. Furthermore, the reduced sulfur is incorporated into cysteine, an amino acid that is a precursor to many other sulfur-containing compounds. In animals, sulfur assimilation occurs primarily through the diet, as animals cannot produce sulfur-containing compounds directly. Sulfur is incorporated into amino acids such as cysteine and methionine, which are used to build proteins and other important molecules.
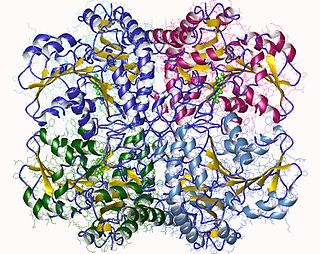
The enzyme cystathionine γ-lyase (EC 4.4.1.1, CTH or CSE; also cystathionase; systematic name L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming)) breaks down cystathionine into cysteine, 2-oxobutanoate (α-ketobutyrate), and ammonia:
Dichloromethane dehalogenase (EC 4.5.1.3; systematic name dichloromethane chloride-lyase (adding H2O; chloride-hydrolysing; formaldehyde-forming)) is a lyase enzyme that generates formaldehyde.
In enzymology, a S-(hydroxymethyl)glutathione dehydrogenase (EC 1.1.1.284) is an enzyme that catalyzes the chemical reaction
In enzymology, a [formate-C-acetyltransferase]-activating enzyme is an enzyme that catalyzes the chemical reaction
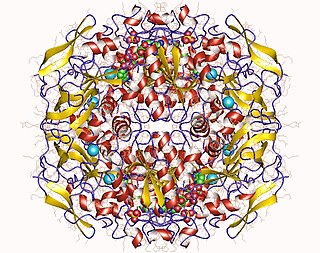
In enzymology, a formaldehyde dehydrogenase (EC 1.2.1.46) is an enzyme that catalyzes the chemical reaction
In enzymology, a mycothiol-dependent formaldehyde dehydrogenase (EC 1.1.1.306) is an enzyme that catalyzes the chemical reaction
The enzyme 3-ketovalidoxylamine C-N-lyase catalyzes the chemical reaction
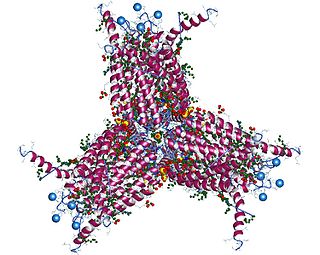
The enzyme leukotriene-C4 synthase (EC 4.4.1.20) catalyzes the reaction
The enzyme 2-dehydropantoate aldolase catalyzes the chemical reaction:
The enzyme dihydroneopterin aldolase catalyzes the chemical reaction
The enzyme dimethylaniline-N-oxide aldolase catalyzes the chemical reaction
The enzyme ketotetrose-phosphate aldolase catalyzes the chemical reaction
The enzyme trimethylamine-oxide aldolase catalyzes the chemical reaction
The enzyme hydroxyacylglutathione hydrolase (EC 3.1.2.6, systematic name = S-(2-hydroxyacyl)glutathione hydrolase) catalyzes the following reaction:
In enzymology, formate C-acetyltransferase is an enzyme. Pyruvate formate lyase is found in Escherichia coli and other organisms. It helps regulate anaerobic glucose metabolism. Using radical non-redox chemistry, it catalyzes the reversible conversion of pyruvate and coenzyme-A into formate and acetyl-CoA. The reaction occurs as follows:

Alcohol dehydrogenase class-3 is an enzyme that in humans is encoded by the ADH5 gene.
The fnr gene of Escherichia coli encodes a transcriptional activator (FNR) which is required for the expression of a number of genes involved in anaerobic respiratory pathways. The FNR protein of E. coli is an oxygen – responsive transcriptional regulator required for the switch from aerobic to anaerobic metabolism.
"Type III mutants, originally frdB, were designated fnr because they were defective in fumarate and nitrate reduction and impaired in their ability to produce gas." - Lambden and Guest, 1976 Journal of General Microbiology97, 145-160







