
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.

Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. Their defining characteristic is their cell envelopes, which consists of a thin peptidoglycan cell wall sandwiched between an inner (cytoplasmic) membrane and an outer membrane. These bacteria are found in all environments that support life on Earth.

Bdellovibrio is a genus of Gram-negative, obligate aerobic bacteria. One of the more notable characteristics of this genus is that members can prey upon other Gram-negative bacteria and feed on the biopolymers, e.g. proteins and nucleic acids, of their hosts. They have two lifestyles: a host-dependent, highly mobile phase, the "attack phase", in which they form "bdelloplasts" in their host bacteria; and a slow-growing, irregularly shaped, host-independent form.

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, from fungal spores (zoospores), and from a wide range of microorganisms to provide motility. Many protists with flagella are known as flagellates.
The evolution of flagella is of great interest to biologists because the three known varieties of flagella – each represent a sophisticated cellular structure that requires the interaction of many different systems.
The cell envelope comprises the inner cell membrane and the cell wall of a bacterium. In gram-negative bacteria an outer membrane is also included. This envelope is not present in the Mollicutes where the cell wall is absent.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.
Members of the genus Selenomonas are referred to trivially as selenomonads. The genus Selenomonas constitutes a group of motile crescent-shaped bacteria and includes species living in the gastrointestinal tracts of animals, in particular the ruminants. A number of smaller forms discovered with the light microscope are now in culture but many, especially the large selenomonads are not, owing to their fastidious and incompletely known growth requirements.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.
The type III secretion system is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. While the type III secretion system has been widely regarded as equivalent to the injectisome, many argue that the injectisome is only part of the type III secretion system, which also include structures like the flagellar export apparatus. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms that evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.
Motility protein A, MotA, is a bacterial protein that is encoded by the motA gene. It is a component of the flagellar motor. More specifically, MotA and MotB make the stator of a H+ driven bacterial flagella and surround the rotor as a ring of about 8–10 particles. MotA and MotB are integral membrane proteins. MotA has four transmembrane domains.
The Negativicutes are a class of bacteria in the phylum Bacillota, whose members have a peculiar cell wall with a lipopolysaccharide outer membrane which stains gram-negative, unlike most other members of the Bacillota. Although several neighbouring Clostridia species also stain gram-negative, the proteins responsible for the unusual diderm structure of the Negativicutes may have actually been laterally acquired from Pseudomonadota. Additional research is required to confirm the origin of the diderm cell envelope in the Negativicutes.
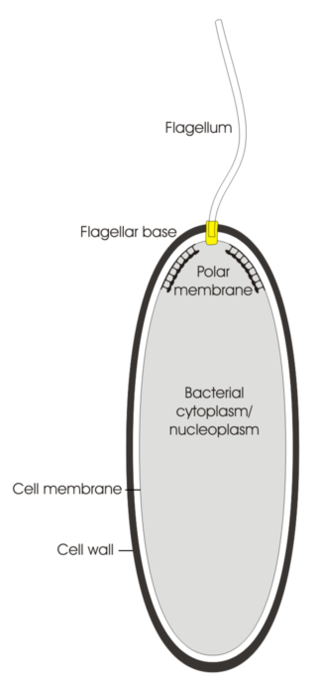
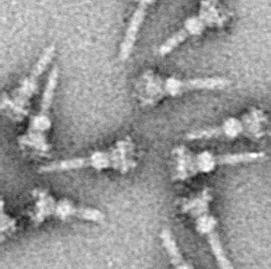
A polar organelle is a structure at a specialised region of the bacterial polar membrane that is associated with the flagellar apparatus. This flagellum-associated structure can easily be distinguished from the other membrane regions in ultrathin sections of embedded bacteria by electron microscopy when the cell membrane is orientated perpendicular to the viewing direction. There, the membrane appears slightly thickened with a finely frilled layer facing the inside of the cell. It is also possible to isolate these polar organelles from the bacterial cells and study them in face view in negatively stained preparations.
Membrane vesicle trafficking in eukaryotic animal cells involves movement of biochemical signal molecules from synthesis-and-packaging locations in the Golgi body to specific release locations on the inside of the plasma membrane of the secretory cell. It takes place in the form of Golgi membrane-bound micro-sized vesicles, termed membrane vesicles (MVs).

Outer membrane vesicles (OMVs) are vesicles released from the outer membranes of Gram-negative bacteria. While Gram-positive bacteria release vesicles as well those vesicles fall under the broader category of bacterial membrane vesicles (MVs). OMVs were the first MVs to be discovered, and are distinguished from outer inner membrane vesicles (OIMVS), which are gram-negative baterial vesicles containing portions of both the outer and inner bacterial membrane. Outer membrane vesicles were first discovered and characterized using transmission-electron microscopy by Indian Scientist Prof. Smriti Narayan Chatterjee and J. Das in 1966-67. OMVs are ascribed the functionality to provide a manner to communicate among themselves, with other microorganisms in their environment and with the host. These vesicles are involved in trafficking bacterial cell signaling biochemicals, which may include DNA, RNA, proteins, endotoxins and allied virulence molecules. This communication happens in microbial cultures in oceans, inside animals, plants and even inside the human body.
Methanogens are a group of microorganisms that produce methane as a byproduct of their metabolism. They play an important role in the digestive system of ruminants. The digestive tract of ruminants contains four major parts: rumen, reticulum, omasum and abomasum. The food with saliva first passes to the rumen for breaking into smaller particles and then moves to the reticulum, where the food is broken into further smaller particles. Any indigestible particles are sent back to the rumen for rechewing. The majority of anaerobic microbes assisting the cellulose breakdown occupy the rumen and initiate the fermentation process. The animal absorbs the fatty acids, vitamins and nutrient content on passing the partially digested food from the rumen to the omasum. This decreases the pH level and initiates the release of enzymes for further breakdown of the food which later passes to the abomasum to absorb remaining nutrients before excretion. This process takes about 9–12 hours.

Twitching motility is a form of crawling bacterial motility used to move over surfaces. Twitching is mediated by the activity of hair-like filaments called type IV pili which extend from the cell's exterior, bind to surrounding solid substrates, and retract, pulling the cell forwards in a manner similar to the action of a grappling hook. The name twitching motility is derived from the characteristic jerky and irregular motions of individual cells when viewed under the microscope. It has been observed in many bacterial species, but is most well studied in Pseudomonas aeruginosa, Neisseria gonorrhoeae and Myxococcus xanthus. Active movement mediated by the twitching system has been shown to be an important component of the pathogenic mechanisms of several species.

Run-and-tumble motion is a movement pattern exhibited by certain bacteria and other microscopic agents. It consists of an alternating sequence of "runs" and "tumbles": during a run, the agent propels itself in a fixed direction, and during a tumble, it remains stationary while it reorients itself in preparation for the next run.
Selenomonas sputigena is a species of anaerobe Gram-negative bacteria that is found in the upper respiratory tract of humans. It is the type species of the genus Selenomonas, with the type strain VPI D 19B-28. It is known to cause blood infection (sepsis), gum inflammation, and tooth decay. It alone cannot damage the tooth enamel, but worsen the damage done by other bacteria such as Streptococcus mutans,Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia.








