
Rhizobia are diazotrophic bacteria that fix nitrogen after becoming established inside the root nodules of legumes (Fabaceae). To express genes for nitrogen fixation, rhizobia require a plant host; they cannot independently fix nitrogen. In general, they are gram negative, motile, non-sporulating rods.

Rhizobium is a genus of Gram-negative soil bacteria that fix nitrogen. Rhizobium species form an endosymbiotic nitrogen-fixing association with roots of (primarily) legumes and other flowering plants.

Ensifer meliloti are an aerobic, Gram-negative, and diazotrophic species of bacteria. S. meliloti are motile and possess a cluster of peritrichous flagella. S. meliloti fix atmospheric nitrogen into ammonia for their legume hosts, such as alfalfa. S. meliloti forms a symbiotic relationship with legumes from the genera Medicago, Melilotus and Trigonella, including the model legume Medicago truncatula. This symbiosis promotes the development of a plant organ, termed a root nodule. Because soil often contains a limited amount of nitrogen for plant use, the symbiotic relationship between S. meliloti and their legume hosts has agricultural applications. These techniques reduce the need for inorganic nitrogenous fertilizers.

Root nodules are found on the roots of plants, primarily legumes, that form a symbiosis with nitrogen-fixing bacteria. Under nitrogen-limiting conditions, capable plants form a symbiotic relationship with a host-specific strain of bacteria known as rhizobia. This process has evolved multiple times within the legumes, as well as in other species found within the Rosid clade. Legume crops include beans, peas, and soybeans.
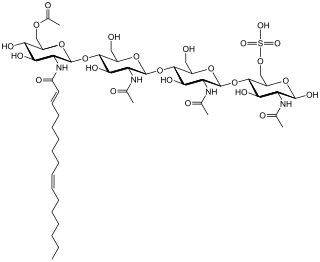
Nod factors, are signaling molecules produced by soil bacteria known as rhizobia in response to flavonoid exudation from plants under nitrogen limited conditions. Nod factors initiate the establishment of a symbiotic relationship between legumes and rhizobia by inducing nodulation. Nod factors produce the differentiation of plant tissue in root hairs into nodules where the bacteria reside and are able to fix nitrogen from the atmosphere for the plant in exchange for photosynthates and the appropriate environment for nitrogen fixation. One of the most important features provided by the plant in this symbiosis is the production of leghemoglobin, which maintains the oxygen concentration low and prevents the inhibition of nitrogenase activity.

N-Acyl homoserine lactones are a class of signaling molecules involved in bacterial quorum sensing, a means of communication between bacteria enabling behaviors based on population density.

Frankia is a genus of nitrogen-fixing bacteria that live in symbiosis with actinorhizal plants, similar to the Rhizobium bacteria found in the root nodules of legumes in the family Fabaceae. Frankia also initiate the forming of root nodules.

Bradyrhizobium is a genus of Gram-negative soil bacteria, many of which fix nitrogen. Nitrogen fixation is an important part of the nitrogen cycle. Plants cannot use atmospheric nitrogen (N2); they must use nitrogen compounds such as nitrates.
Ensifer is a genus of nitrogen-fixing bacteria (rhizobia), three of which have been sequenced.
The nif genes are genes encoding enzymes involved in the fixation of atmospheric nitrogen into a form of nitrogen available to living organisms. The primary enzyme encoded by the nif genes is the nitrogenase complex which is in charge of converting atmospheric nitrogen (N2) to other nitrogen forms such as ammonia which the organism can use for various purposes. Besides the nitrogenase enzyme, the nif genes also encode a number of regulatory proteins involved in nitrogen fixation. The nif genes are found in both free-living nitrogen-fixing bacteria and in symbiotic bacteria associated with various plants. The expression of the nif genes is induced as a response to low concentrations of fixed nitrogen and oxygen concentrations (the low oxygen concentrations are actively maintained in the root environment of host plants). The first Rhizobium genes for nitrogen fixation (nif) and for nodulation (nod) were cloned in the early 1980s by Gary Ruvkun and Sharon R. Long in Frederick M. Ausubel's laboratory.

Within genetics, post-genomic research has rendered bacterial small non-coding RNAs (sRNAs) as major players in post-transcriptional regulation of gene expression in response to environmental stimuli. The Alphaproteobacteria includes Gram-negative microorganisms with diverse life styles; frequently involving long-term interactions with higher eukaryotes.
αr7 is a family of bacterial small non-coding RNAs with representatives in a broad group of Alphaproteobacterial species from the order Hyphomicrobiales. The first member of this family was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis identified full-length homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr7 RNA species are 134-159 nucleotides (nt) long and share a well defined common secondary structure. αr7 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the Alphaproteobacterial genomes.
αr14 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria. The first member of this family (Smr14C2) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). It was later renamed NfeR1 and shown to be highly expressed in salt stress and during the symbiotic interaction on legume roots. Further homology and structure conservation analysis identified 2 other chromosomal copies and 3 plasmidic ones. Moreover, full-length Smr14C homologs have been identified in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr14C RNA species are 115-125 nt long and share a well defined common secondary structure. Most of the αr14 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr15 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first members of this family were found tandemly arranged in the same intergenic region (IGR) of the Sinorhizobium meliloti 1021 chromosome (C). Further homology and structure conservation analysis have identified full-length Smr15C1 and Smr15C2 homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. The Smr15C1 and Smr15C2 homologs are also encoded in tandem within the same IGR region of Rhizobium and Agrobacterium species, whereas in Brucella species the αr15C loci are spread in the IGRs of Chromosome I. Moreover, this analysis also identified a third αr15 loci in extrachromosomal replicons of the mentioned nitrogen-fixing α-proteobacteria and in the Chromosome II of Brucella species. αr15 RNA species are 99-121 nt long and share a well defined common secondary structure consisting of three stem loops. The transcripts of the αr15 family can be catalogued as trans-acting sRNAs encoded by independent transcription units with recognizable promoter and transcription termination signatures within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr35 is a family of bacterial small non-coding RNAs with representatives in a reduced group of Alphaproteobacteria from the order Hyphomicrobiales. The first member of this family (Smr35B) was found in a Sinorhizobium meliloti 1021 locus located in the symbiotic plasmid B (pSymB). Further homology and structure conservation analysis have identified full-length SmrB35 homologs in other legume symbionts, as well as in the human and plant pathogens Brucella anthropi and Agrobacterium tumefaciens, respectively. αr35 RNA species are 139-142 nt long and share a common secondary structure consisting of two stem loops and a well conserved rho independent terminator. Most of the αr35 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions of the Alphaproteobacterial genomes.
αr45 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Hyphomicrobiales. The first member of this family (Smr45C) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis identified homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species, in Bartonella species, in several members of the Xanthobactereacea family, and in some representatives of the Beijerinckiaceae family. αr45C RNA species are 147-153 nt long and share a well defined common secondary structure. All of the αr45 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
Mesorhizobium loti, formerly known as Rhizobium loti, is a Gram negative species of bacteria found in the root nodules of many plant species. Its name is a reference to Lotus corniculatus, a flowering plant from which it was originally isolated.
Mesorhizobium mediterraneum is a bacterium from the genus Mesorhizobium, which was isolated from root nodule of the Chickpea in Spain. The species Rhizobium mediterraneum was subsequently transferred to Mesorhizobium mediterraneum. This species, along with many other closely related taxa, have been found to promote production of chickpea and other crops worldwide by forming symbiotic relationships.
Ensifer medicae is a species of gram-negative, nitrogen-fixing, rod-shaped bacteria. They can be free-living or symbionts of leguminous plants in root nodules. E.medicae was first isolated from root nodules on plants in the genus Medicago. Some strains of E.medicae, like WSM419, are aerobic. They are chemoorganotrophic mesophiles that prefer temperatures around 28 °C. In addition to their primary genome, these organisms also have three known plasmids, sized 1,570,951 bp, 1,245,408 bp and 219,313 bp.

A symbiosome is a specialised compartment in a host cell that houses an endosymbiont in a symbiotic relationship.









