Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.

Chlorophyll b is a form of chlorophyll. Chlorophyll b helps in photosynthesis by absorbing light energy. It is more soluble than chlorophyll a in polar solvents because of its carbonyl group. Its color is green, and it primarily absorbs blue light.

Protoporphyrin ferrochelatase (EC 4.98.1.1, formerly EC 4.99.1.1, or ferrochelatase; systematic name protoheme ferro-lyase (protoporphyrin-forming)) is an enzyme encoded by the FECH gene in humans. Ferrochelatase catalyses the eighth and terminal step in the biosynthesis of heme, converting protoporphyrin IX into heme B. It catalyses the reaction:

The transsulfuration pathway is a metabolic pathway involving the interconversion of cysteine and homocysteine through the intermediate cystathionine. Two transsulfurylation pathways are known: the forward and the reverse.

In enzymology, a precorrin-2 dehydrogenase (EC 1.3.1.76) is an enzyme that catalyzes the chemical reaction

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.
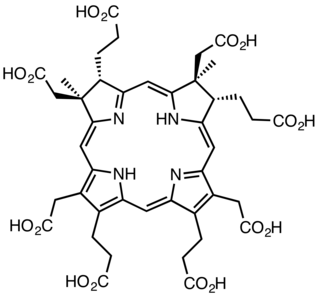
The enzyme sirohydrochlorin cobaltochelatase (EC 4.99.1.3) catalyzes the reaction
The enzyme threonine-phosphate decarboxylase (EC 4.1.1.81) catalyzes the chemical reaction

Cobalt chelatase (EC 6.6.1.2) is an enzyme that catalyzes the chemical reaction
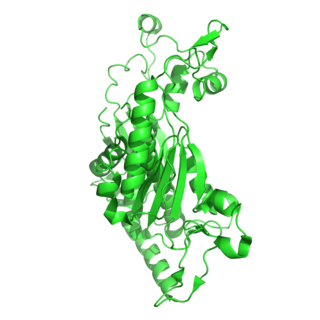
The enzyme chorismate synthase catalyzes the chemical reaction

In enzymology, a homocitrate synthase (EC 2.3.3.14) is an enzyme that catalyzes the chemical reaction

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.
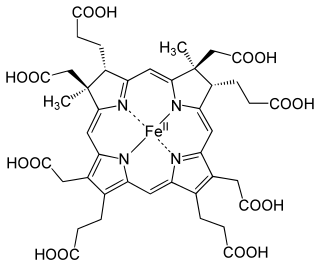
Siroheme is a heme-like prosthetic group at the active sites of some enzymes to accomplish the six-electron reduction of sulfur and nitrogen. It is a cofactor at the active site of sulfite reductase, which plays a major role in sulfur assimilation pathway, converting sulfite into sulfide, which can be incorporated into the organic compound homocysteine.

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.
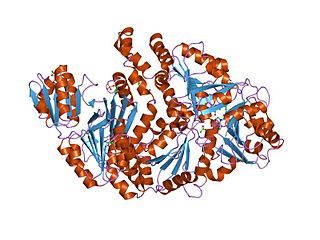
In molecular biology, sirohaem synthase (CysG) is a multi-functional enzyme with S-adenosyl-L-methionine (SAM)-dependent bismethyltransferase, dehydrogenase and ferrochelatase activities. Bacterial sulphur metabolism depends on the iron-containing porphinoid sirohaem. CysG synthesizes sirohaem from uroporphyrinogen III via reactions which encompass two branchpoint intermediates in tetrapyrrole biosynthesis, diverting flux first from protoporphyrin IX biosynthesis and then from cobalamin biosynthesis. CysG is a dimer. Its dimerisation region is 74 amino acids long, and acts to hold the two structurally similar protomers held together asymmetrically through a number of salt-bridges across complementary residues within the dimerisation region. CysG dimerisation produces a series of active sites, accounting for CysG's multi-functionality, catalysing four diverse reactions:

Ferredoxin-thioredoxin reductase EC 1.8.7.2, systematic name ferredoxin:thioredoxin disulfide oxidoreductase, is a [4Fe-4S] protein that plays an important role in the ferredoxin/thioredoxin regulatory chain. It catalyzes the following reaction:

F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR). MCR catalyzes the reaction EC 2.8.4.1 that releases methane in the final step of methanogenesis:
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.

Uroporphyrinogen-III C-methyltransferase, uroporphyrinogen methyltransferase, uroporphyrinogen-III methyltransferase, adenosylmethionine-uroporphyrinogen III methyltransferase, S-adenosyl-L-methionine-dependent uroporphyrinogen III methylase, uroporphyrinogen-III methylase, SirA, CysG, CobA, uroporphyrin-III C-methyltransferase, S-adenosyl-L-methionine:uroporphyrin-III C-methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:uroporphyrinogen-III C-methyltransferase. This enzyme catalyses the following chemical reaction

















