Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.

Homoserine (also called isothreonine) is an α-amino acid with the chemical formula HO2CCH(NH2)CH2CH2OH. L-Homoserine is not one of the common amino acids encoded by DNA. It differs from the proteinogenic amino acid serine by insertion of an additional -CH2- unit into the backbone. Homoserine, or its lactone form, is the product of a cyanogen bromide cleavage of a peptide by degradation of methionine.
In enzymology, a succinylglutamate-semialdehyde dehydrogenase (EC 1.2.1.71) is an enzyme that catalyzes the chemical reaction
In enzymology, a N-succinylarginine dihydrolase (EC 3.5.3.23) is an enzyme that catalyzes the chemical reaction
In enzymology, a succinylglutamate desuccinylase (EC 3.5.1.96) is an enzyme that catalyzes the chemical reaction
In enzymology, an arginine N-succinyltransferase (EC 2.3.1.109) is an enzyme that catalyzes the chemical reaction

In enzymology, 4-aminobutyrate transaminase, also called GABA transaminase or 4-aminobutyrate aminotransferase, or GABA-T, is an enzyme that catalyzes the chemical reaction:
In enzymology, an acetylornithine transaminase (EC 2.6.1.11) is an enzyme that catalyzes the chemical reaction
In enzymology, an arginine-pyruvate transaminase is an enzyme that catalyzes the chemical reaction
In enzymology, a D-amino-acid transaminase is an enzyme that catalyzes the chemical reaction:
In enzymology, a diaminobutyrate-2-oxoglutarate transaminase is an enzyme that catalyzes the chemical reaction

In enzymology, a kynurenine-oxoglutarate transaminase is an enzyme that catalyzes the chemical reaction
In enzymology, a L-lysine 6-transaminase is an enzyme that catalyzes the chemical reaction
In enzymology, a N6-acetyl-beta-lysine transaminase is an enzyme that catalyzes the chemical reaction
In enzymology, an ornithine(lysine) transaminase (EC 2.6.1.68) is an enzyme that catalyzes the chemical reaction
In enzymology, a succinyldiaminopimelate transaminase is an enzyme that catalyzes the chemical reaction
In enzymology, a taurine-2-oxoglutarate transaminase is an enzyme that catalyzes the chemical reaction.
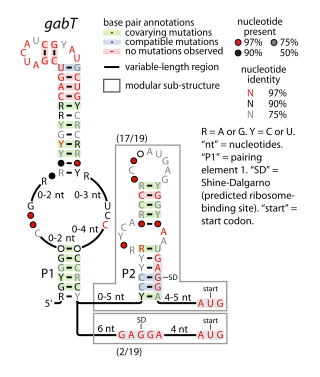
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gab operon is responsible for the conversion of γ-aminobutyrate (GABA) to succinate. The gab operon comprises three structural genes – gabD, gabT and gabP – that encode for a succinate semialdehyde dehydrogenase, GABA transaminase and a GABA permease respectively. There is a regulatory gene csiR, downstream of the operon, that codes for a putative transcriptional repressor and is activated when nitrogen is limiting.
N-succinylornithine carbamoyltransferase (EC 2.1.3.11, succinylornithine transcarbamylase, N-succinyl-L-ornithine transcarbamylase, SOTCase) is an enzyme with systematic name carbamoyl phosphate:N2-succinyl-L-ornithine carbamoyltransferase. This enzyme catalyses the following chemical reaction






