
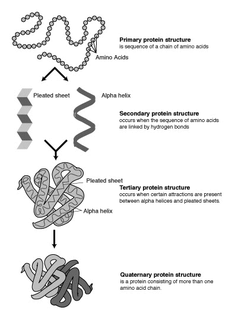
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence.

The beta sheet, (β-sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease.

Protein secondary structure is the three dimensional form of local segments of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine and biotechnology.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA.

The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold. The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.

Cyclic nucleotide–gated ion channels or CNG channels are ion channels that function in response to the binding of cyclic nucleotides. CNG channels are nonselective cation channels that are found in the membranes of various tissue and cell types, and are significant in sensory transduction as well as cellular development. Their function can be the result of a combination of the binding of cyclic nucleotides and either a depolarization or a hyperpolarization event. Initially discovered in the cells that make up the retina of the eye, CNG channels have been found in many different cell types across both the animal and the plant kingdoms. CNG channels have a very complex structure with various subunits and domains that play a critical role in their function. CNG channels are significant in the function of various sensory pathways including vision and olfaction, as well as in other key cellular functions such as hormone release and chemotaxis. CNG channels have also been found to exist in prokaryotes, including many spirochaeta, though their precise role in bacterial physiology remains unknown.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
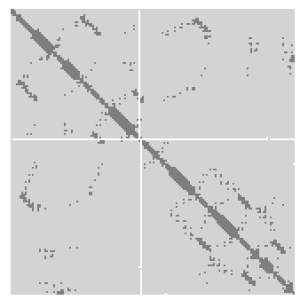
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional matrix. For two residues and , the element of the matrix is 1 if the two residues are closer than a predetermined threshold, and 0 otherwise. Various contact definitions have been proposed: The distance between the Cα-Cα atom with threshold 6-12 Å; distance between Cβ-Cβ atoms with threshold 6-12 Å ; and distance between the side-chain centers of mass.

In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function contain the beta barrel structure.
A turn is an element of secondary structure in proteins where the polypeptide chain reverses its overall direction.
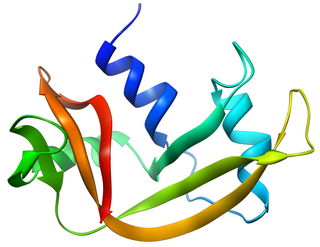
Bovine pancreatic ribonuclease, also often referred to as bovine pancreatic ribonuclease A or simply RNase A, is a pancreatic ribonuclease enzyme that cleaves single-stranded RNA. Bovine pancreatic ribonuclease is one of the classic model systems of protein science. Two Nobel Prizes in Chemistry have been awarded in recognition of work on bovine pancreatic ribonuclease: in 1972, the Prize was awarded to Christian Anfinsen for his work on protein folding and to Stanford Moore and William Stein for their work on the relationship between the protein's structure and its chemical mechanism; in 1984, the Prize was awarded to Robert Bruce Merrifield for development of chemical synthesis of proteins.
A beta bulge can be described as a localized disruption of the regular hydrogen bonding of beta sheet by inserting extra residues into one or both hydrogen bonded β-strands.
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.

The beta hairpin is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction, and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.
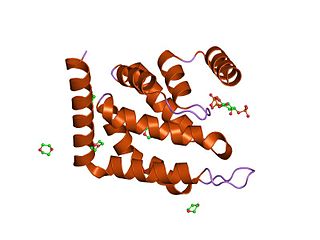
The epsin N-terminal homology (ENTH) domain is a structural domain that is found in proteins involved in endocytosis and cytoskeletal machinery.
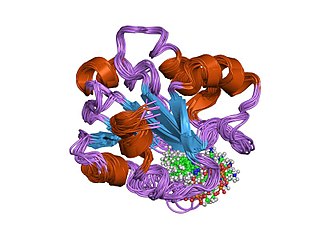
In molecular biology, the vitamin B12-binding domain is a protein domain which binds to cobalamin. It can bind two different forms of the cobalamin cofactor, with cobalt bonded either to a methyl group (methylcobalamin) or to 5'-deoxyadenosine (adenosylcobalamin). Cobalamin-binding domains are mainly found in two families of enzymes present in animals and prokaryotes, which perform distinct kinds of reactions at the cobalt-carbon bond. Enzymes that require methylcobalamin carry out methyl transfer reactions. Enzymes that require adenosylcobalamin catalyse reactions in which the first step is the cleavage of adenosylcobalamin to form cob(II)alamin and the 5'-deoxyadenosyl radical, and thus act as radical generators. In both types of enzymes the B12-binding domain uses a histidine to bind the cobalt atom of cobalamin cofactors. This histidine is embedded in a DXHXXG sequence, the most conserved primary sequence motif of the domain. Proteins containing the cobalamin-binding domain include:
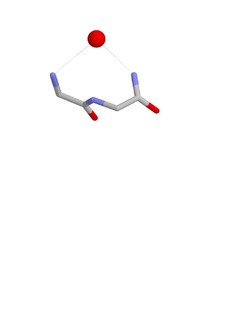
The Nest is a type of protein structural motif. It is a small recurring anion-binding feature of both proteins and peptides. Each consists of the main chain atoms of three consecutive amino acid residues. The main chain NH groups bind the anions while the side chain atoms are often not involved. Proline residues lack NH groups so are rare in nests. About one in 12 of amino acid residues in proteins, on average, belongs to a nest.


















