
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.

Meiosis is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately result in four cells with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and female will fuse to create a cell with two copies of each chromosome again, the zygote.

Condensins are large protein complexes that play a central role in chromosome assembly and segregation during mitosis and meiosis. Their subunits were originally identified as major components of mitotic chromosomes assembled in Xenopus egg extracts.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.

Separase, also known as separin, is a cysteine protease responsible for triggering anaphase by hydrolysing cohesin, which is the protein responsible for binding sister chromatids during the early stage of anaphase. In humans, separin is encoded by the ESPL1 gene.

A chromomere, also known as an idiomere, is one of the serially aligned beads or granules of a eukaryotic chromosome, resulting from local coiling of a continuous DNA thread. Chromeres are regions of chromatin that have been compacted through localized contraction. In areas of chromatin with the absence of transcription, condensing of DNA and protein complexes will result in the formation of chromomeres. It is visible on a chromosome during the prophase of meiosis and mitosis. Giant banded (Polytene) chromosomes resulting from the replication of the chromosomes and the synapsis of homologs without cell division is a process called endomitosis. These chromosomes consist of more than 1000 copies of the same chromatid that are aligned and produce alternating dark and light bands when stained. The dark bands are the chromomere.
SMC complexes represent a large family of ATPases that participate in many aspects of higher-order chromosome organization and dynamics. SMC stands for Structural Maintenance of Chromosomes.
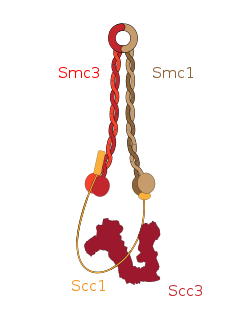
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination, and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.

Structural maintenance of chromosomes protein 1A (SMC1A) is a protein that in humans is encoded by the SMC1A gene. SMC1A is a subunit of the cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. In somatic cells, cohesin is formed of SMC1A, SMC3, RAD21 and either SA1 or SA2 whereas in meiosis, cohesin is formed of SMC3, SMC1B, REC8 and SA3.

Double-strand-break repair protein rad21 homolog is a protein that in humans is encoded by the RAD21 gene. RAD21, an essential gene, encodes a DNA double-strand break (DSB) repair protein that is evolutionarily conserved in all eukaryotes from budding yeast to humans. RAD21 protein is a structural component of the highly conserved cohesin complex consisting of RAD21, SMC1A, SMC3, and SCC3 [ STAG1 (SA1) and STAG2 (SA2) in multicellular organisms] proteins, involved in sister chromatid cohesion.

Structural maintenance of chromosomes protein 3 (SMC3) is a protein that in humans is encoded by the SMC3 gene. SMC3 is a subunit of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, RAD21 and either SA1 or SA2. In humans, SMC3 is present in all cohesin complexes whereas there are multiple paralogs for the other subunits.

Cohesin subunit SA-2 (SA2) is a protein that in humans is encoded by the STAG2 gene. SA2 is a subunit of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. In somatic cells cohesin is formed of SMC3, SMC1, RAD21 and either SA1 or SA2 whereas in meiosis, cohesin is formed of SMC3, SMC1B, REC8 and SA3.

Shugoshin-like 1 is a protein that in humans is encoded by the SGOL1 gene.

Sister chromatid cohesion protein PDS5 homolog B(PDS5B) is a protein that in humans is encoded by the PDS5B gene. It is a regulatory subunit of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. The core cohesin complex is formed of SMC3, SMC1, RAD21 and either SA1 or SA2. PDS5 associates with WAPL to stimulate the release of cohesin from DNA but during DNA replication PDS5 promotes acetylation of SMC3 by ESCO1 and ESCO2.
Structural maintenance of chromosomes protein 5 is a protein encoded by the SMC5 gene in human.

Sister chromatid cohesion protein PDS5 homolog A is a protein that in humans is encoded by the PDS5A gene.

Meiotic recombination protein REC8 homolog is a protein that in humans is encoded by the REC8 gene.
Sister chromatid cohesion refers to the process by which sister chromatids are paired and held together during certain phases of the cell cycle. Establishment of sister chromatid cohesion is the process by which chromatin-associated cohesin protein becomes competent to physically bind together the sister chromatids. In general, cohesion is established during S phase as DNA is replicated, and is lost when chromosomes segregate during mitosis and meiosis. Some studies have suggested that cohesion aids in aligning the kinetochores during mitosis by forcing the kinetochores to face opposite cell poles.

Structural maintenance of chromosomes protein 1B (SMC-1B) is a protein that in humans is encoded by the SMC1B gene. SMC-1B belongs to a family of proteins required for chromatid cohesion and DNA recombination during meiosis and mitosis. SMC1ß protein appears to participate with other cohesins REC8, STAG3 and SMC3 in sister-chromatid cohesion throughout the whole meiotic process in human oocytes.























