Related Research Articles
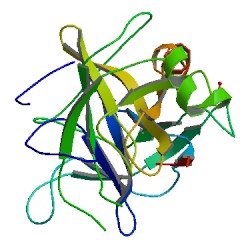
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
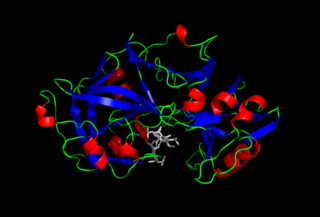
Pepsin is an endopeptidase that breaks down proteins into smaller peptides. It is produced in the gastric chief cells of the stomach lining and is one of the main digestive enzymes in the digestive systems of humans and many other animals, where it helps digest the proteins in food. Pepsin is an aspartic protease, using a catalytic aspartate in its active site.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Neurophysin I is a carrier protein with a size of 10 KDa and contains 90 to 97 amino acids. It is a cleavage product of preprooxyphysin. It is a neurohypophysial hormone that is transported in vesicles with oxytocin, the other cleavage product, along axons, from magnocellular neurons of the hypothalamus to the posterior lobe of the pituitary. Although it is stored in neurosecretory granules with oxytocin and released with oxytocin, its biological action is unclear.

Furin is a protease, a proteolytic enzyme that in humans and other animals is encoded by the FURIN gene. Some proteins are inactive when they are first synthesized, and must have sections removed in order to become active. Furin cleaves these sections and activates the proteins. It was named furin because it was in the upstream region of an oncogene known as FES. The gene was known as FUR and therefore the protein was named furin. Furin is also known as PACE. A member of family S8, furin is a subtilisin-like peptidase.
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.

HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious.
Many major physiological processes depend on regulation of proteolytic enzyme activity and there can be dramatic consequences when equilibrium between an enzyme and its substrates is disturbed. In this prospective, the discovery of small-molecule ligands, like protease inhibitors, that can modulate catalytic activities has an enormous therapeutic effect. Hence, inhibition of the HIV protease is one of the most important approaches for the therapeutic intervention in HIV infection and their development is regarded as major success of structure-based drug design. They are highly effective against HIV and have, since the 1990s, been a key component of anti-retroviral therapies for HIV/AIDS.
Hepsin is an enzyme. This enzyme catalyses the following chemical reaction
Kallikrein 8 is an enzyme. This enzyme catalyses the following chemical reaction
Pro-opiomelanocortin converting enzyme is an enzyme. This enzyme catalyses the following chemical reaction
Mucorpepsin is an enzyme. This enzyme catalyses the following chemical reaction

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.
Magnolysin is an enzyme. This enzyme catalyses the following chemical reaction

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.

Cinnamycin is a tetracyclic antibacterial peptide produced by Streptomyces cinnamoneus containing 19 amino acid residues including the unusual amino acids threo-3-methyl-lanthionine, meso-lanthionine, lysinoalanine, and 3-hydroxyaspartic acid.
References
- ↑ Cawley NX, Chen HC, Beinfeld MC, Loh YP (February 1996). "Specificity and kinetic studies on the cleavage of various prohormone mono- and paired-basic residue sites by yeast aspartic protease 3". The Journal of Biological Chemistry. 271 (8): 4168–76. doi: 10.1074/jbc.271.8.4168 . PMID 8626758.
- ↑ Fuller RS, Rawlings ND, Woessner JF (1998). "Yapsin 2". In Barrett AJ (ed.). Handbook of Proteolytic Enzymes. London: Academic Press. pp. 908–909.
- ↑ Olsen V, Guruprasad K, Cawley NX, Chen HC, Blundell TL, Loh YP (March 1998). "Cleavage efficiency of the novel aspartic protease yapsin 1 (Yap3p) enhanced for substrates with arginine residues flanking the P1 site: correlation with electronegative active-site pockets predicted by molecular modeling". Biochemistry. 37 (9): 2768–77. doi:10.1021/bi9724826. PMID 9485427.