| |
| Names | |
|---|---|
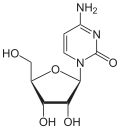
| IUPAC name 1-(β-D-Ribofuranosyl)pyrimidin-2(1H)-one | |
| Systematic IUPAC name 1-[(2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2(1H)-one | |
| Other names Pyrimidin-2-one β-D-ribofuranoside | |
| Identifiers | |
3D model (JSmol) | |
| ChemSpider | |
PubChem CID | |
| UNII | |
CompTox Dashboard (EPA) | |
| |
| |
| Properties | |
| C9H12N2O5 | |
| Molar mass | 228.204 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Zebularine is a nucleoside analog of cytidine. It is a transition state analog inhibitor of cytidine deaminase by binding to the active site as covalent hydrates. Also shown to inhibit DNA methylation and tumor growth both in vitro and in vivo. [1]
In a small study of mice with a defective Adenomatous polyposis coli gene, oral administration of zebularine to males had no effect on the overall methylation of DNA or the number of polyps, but in females the average number of polyps was reduced from 58 to 1. [2] It has therefore been suggested for drug use as a prototype of epigenetic therapy for cancer chemoprevention. [3]