Related Research Articles
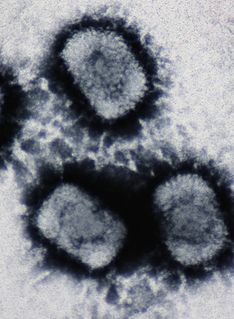
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: Duplodnaviria and Varidnaviria, and ssDNA viruses are almost exclusively assigned to the realm Monodnaviria, which also includes dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Viruses that have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase are separately considered reverse transcribing viruses and are assigned to the kingdom Pararnavirae in the realm Riboviria.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.
An oncolytic virus is a virus that preferentially infects and kills cancer cells. As the infected cancer cells are destroyed by oncolysis, they release new infectious virus particles or virions to help destroy the remaining tumour. Oncolytic viruses are thought not only to cause direct destruction of the tumour cells, but also to stimulate host anti-tumour immune system responses.

Virus latency is the ability of a pathogenic virus to lie dormant (latent) within a cell, denoted as the lysogenic part of the viral life cycle. A latent viral infection is a type of persistent viral infection which is distinguished from a chronic viral infection. Latency is the phase in certain viruses' life cycles in which, after initial infection, proliferation of virus particles ceases. However, the viral genome is not eradicated. The virus can reactivate and begin producing large amounts of viral progeny without the host becoming reinfected by new outside virus, and stays within the host indefinitely.
HHV Latency Associated Transcript is a length of RNA which accumulates in cells hosting long-term, or latent, Human Herpes Virus (HHV) infections. The LAT RNA is produced by genetic transcription from a certain region of the viral DNA. LAT regulates the viral genome and interferes with the normal activities of the infected host cell.

Herpesviridae is a large family of DNA viruses that cause infections and certain diseases in animals, including humans. The members of this family are also known as herpesviruses. The family name is derived from the Greek word herpein, referring to spreading cutaneous lesions, usually involving blisters, seen in flares of herpes simplex 1, herpes simplex 2 and herpes zoster (shingles). In 1971, the International Committee on the Taxonomy of Viruses (ICTV) established Herpesvirus as a genus with 23 viruses among four groups. Currently, 107 species are recognized, all but one of which are in one of the three subfamilies.

Human Herpes Virus (HHV) Infected Cell Polypeptide 0 (ICP0) is a protein, encoded by the DNA of herpes viruses. It is produced by herpes viruses during the earliest stage of infection, when the virus has recently entered the host cell; this stage is known as the immediate-early or α ("alpha") phase of viral gene expression. During these early stages of infection, ICP0 protein is synthesized and transported to the nucleus of the infected host cell. Here, ICP0 promotes transcription from viral genes, disrupts structures in the nucleus known as nuclear dots or promyelocytic leukemia (PML) nuclear bodies, and alters the expression of host and viral genes in combination with a neuron specific protein. At later stages of cellular infection, ICP0 relocates to the cell cytoplasm to be incorporated into new virion particles.

Herpes simplex virus1 and 2, also known by their taxonomical names Human alphaherpesvirus 1 and Human alphaherpesvirus 2, are two members of the human Herpesviridae family, a set of new viruses that produce viral infections in the majority of humans. Both HSV-1 and HSV-2 are common and contagious. They can be spread when an infected person begins shedding the virus.

RNA-dependent RNA polymerase or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyses synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
Anthony (Tony) Charles Minson, PhD, FMedSci is a British virologist known for his work on the biology of herpesviruses, and a university administrator. He was the Senior Pro-Vice-Chancellor of the University of Cambridge from 2003 to 2009. He is emeritus professor of virology at the university's Department of Pathology and an emeritus fellow of Wolfson College.

Herpesvirus glycoprotein B is a viral glycoprotein that is involved in the viral cell entry of Herpes simplex virus (HSV). Herpesviruses have an envelope and an outer lipid bilayer which contains twelve surface glycoproteins. For infectivity to be attained, the double stranded DNA genome of HSV must enter the host cell through means of fusion of its envelope with the cellular membrane or via endocytosis. Other viral glycoproteins involved in the process of viral cell entry include gC, gB, gD, gH, and gL, but only gC, gB, gD, and gH are required for the fusion of the HSV's envelope with the cellular membrane. It can be noted that all herpesviruses have glycoproteins gB, gH, and gL.
Herpes simplex research includes all medical research that attempts to prevent, treat, or cure herpes, as well as fundamental research about the nature of herpes. Examples of particular herpes research include drug development, vaccines and genome editing. HSV-1 and HSV-2 are commonly thought of as oral and genital herpes respectively, but other members in the herpes family include chickenpox (varicella/zoster), cytomegalovirus, and Epstein-Barr virus. There are many more members that infect animals other than humans, some of which cause disease in companion animals or have economic impacts in the agriculture industry.

Many variants of herpes simplex virus have been considered for viral therapy of cancer; the early development of these was thoroughly reviewed in the journal Cancer Gene Therapy in 2002. This page describes the most notable variants—those tested in clinical trials: G207, HSV1716, NV1020 and Talimogene laherparepvec. These attenuated versions are constructed by deleting viral genes required for infecting or replicating inside normal cells but not cancer cells, such as ICP34.5, ICP6/UL39, and ICP47.
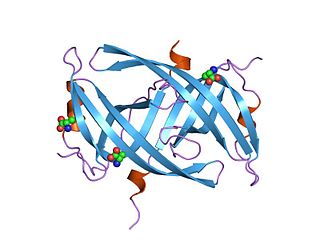
Single-stranded binding proteins (SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans.
David M. Knipe is the Higgins Professor of Microbiology and Molecular Genetics and interim Co-Chair in the Department of Microbiology and Immunobiology at the Harvard Medical School in Boston, Massachusetts and co-chief editor of the reference book Fields Virology. He had previously served as Chair of the Program in Virology at Harvard Medical School from 2004 through 2016.
Human Herpes Viruses, also known as HHVs, are part of a family of DNA viruses that cause several diseases in humans. One of the most notable functions of this virus family is their ability to enter a latent phase and lay dormant within animals for extended periods of time. The mechanism that controls this is very complex because expression of viral proteins during latency is decreased a great deal, meaning that the virus must have transcription of its genes repressed. There are many factors and mechanisms that control this process and epigenetics is one way this is accomplished. Epigenetics refers to persistent changes in expression patterns that are not caused by changes to the DNA sequence. This happens through mechanisms such as methylation and acetylation of histones, DNA methylation, and non-coding RNAs (ncRNA). Altering the acetylation of histones creates changes in expression by changing the binding affinity of histones to DNA, making it harder or easier for transcription machinery to access the DNA. Methyl and acetyl groups can also act as binding sites for transcription factors and enzymes that further modify histones or alter the DNA itself.
Infected cell protein 34.5 is a protein expressed by the ɣ34.5 gene in viruses such as herpes simplex virus; it blocks a cellular stress response to viral infection. It shares the C-terminal regulatory domain with protein phosphatase 1 subunit 15A/B.
HSV epigenetics is the epigenetic modification of herpes simplex virus (HSV) genetic code.
In virology, realm is the highest taxonomic rank established for viruses by the International Committee on Taxonomy of Viruses (ICTV), which oversees virus taxonomy. Four virus realms are recognized and united by specific highly conserved traits: Duplodnaviria, which contains all double-stranded DNA (dsDNA) viruses that encode the HK97-fold major capsid protein, Monodnaviria, which contains all single-stranded DNA (ssDNA) viruses that encode a HUH superfamily endonuclease and their descendents, Riboviria, which contains all RNA viruses that encode RNA-dependent RNA polymerase and all viruses that encode reverse transcriptase, and Varidnaviria, which contains all dsDNA viruses that encode a vertical jelly roll major capsid protein.
References
- 1 2 3 Boehmer, PE; Lehman, IR (1993). "Herpes simplex virus type 1 ICP8: Helix-destabilizing properties". Journal of Virology. 67 (2): 711–5. PMC 237422 . PMID 8380461.
- ↑ Zhou, C; Knipe, DM (2002). "Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme". Journal of Virology. 76 (12): 5893–904. doi:10.1128/JVI.76.12.5893-5904.2002. PMC 136207 . PMID 12021322.