Function
Osteonectin is an acidic extracellular matrix glycoprotein that plays a vital role in bone mineralization, cell-matrix interactions, and collagen binding. Osteonectin also increases the production and activity of matrix metalloproteinases, a function important to invading cancer cells within bone. Additional functions of osteonectin beneficial to tumor cells include angiogenesis, proliferation and migration. Overexpression of osteonectin is reported in many human cancers such as breast, prostate, colon and pancreatic. [6]
This molecule has been implicated in several biological functions, including mineralization of bone and cartilage, inhibiting mineralization, modulation of cell proliferation, facilitation of acquisition of differentiated phenotype and promotion of cell attachment and spreading.
A number of phosphoproteins and glycoproteins are found in bone. The phosphate is bound to the protein backbone through phosphorylated serine or threonine amino acid residues. The best characterized of these bone proteins is osteonectin. It binds collagen and hydroxyapatite in separate domains, is found in relatively large amounts in immature bone, and promotes mineralization of collagen.
Tissue distribution
Fibroblasts, including periodontal fibroblasts, synthesize osteonectin. [7] This protein is synthesized by macrophages at sites of wound repair and platelet degranulation, so it may play an important role in wound healing. SPARC does not support cell attachment, and like tenascin, is anti-adhesive and an inhibitor of cell spreading. It disrupts focal adhesions in fibroblasts. It also regulates the proliferation of some cells, especially endothelial cells, mediated by its ability to bind to cytokines and growth factors. [8] Osteonectin has also been found to decrease DNA synthesis in cultured bone. [9]
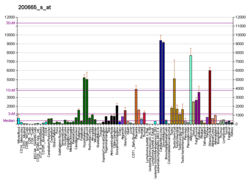
High levels of immunodetectable osteonectin are found in active osteoblasts and marrow progenitor cells, odontoblasts, periodontal ligament and gingival cells, and some chondrocytes and hypertrophic chondrocytes. Osteonectin is also detectable in osteoid, bone matrix proper, and dentin. Osteonectin has been localized in a variety of tissues, but is found in greatest abundance in osseous tissue, tissues characterized by high turnover (such as intestinal epithelium), basement membranes, and certain neoplasms. Osteonectin is expressed by a wide variety of cells, including chondrocytes, fibroblasts, platelets, endothelial cells, epithelial cells, Leydig cells, Sertoli cells, luteal cells, adrenal cortical cells, and numerous neoplastic cell lines (such as SaOS-2 cells from human osteosarcoma).
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.