IQ motif containing GTPase activating protein (IQGAP) is a carrier protein.
Contents
It is associated with the Rho GTP-binding protein.
IQ motif containing GTPase activating protein (IQGAP) is a carrier protein.
It is associated with the Rho GTP-binding protein.
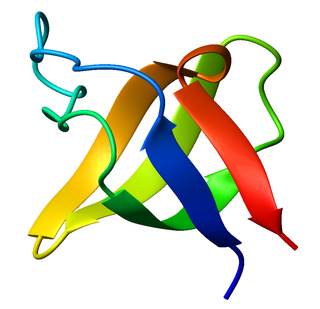
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.
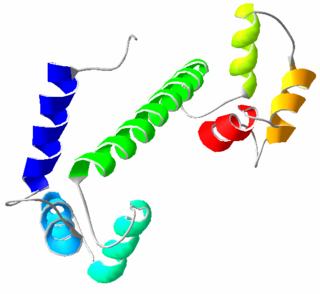
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) which stabilizes the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an N-glycosylation site motif can be defined as Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue.

The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold. The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.

Helix-turn-helix is a DNA-binding domain (DBD). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.
Calmodulin-binding proteins are, as their name implies, proteins which bind calmodulin. Calmodulin can bind to a variety of proteins through a two-step binding mechanism, namely "conformational and mutually induced fit", where typically two domains of calmodulin wrap around an emerging helical calmodulin binding domain from the target protein.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.

Ras GTPase-activating-like protein IQGAP1 (IQGAP1) also known as p195 is a ubiquitously expressed protein that in humans is encoded by the IQGAP1 gene. IQGAP1 is a scaffold protein involved in regulating various cellular processes ranging from organization of the actin cytoskeleton, transcription, and cellular adhesion to regulating the cell cycle.

Calcium binding protein 1 is a protein that in humans is encoded by the CABP1 gene. Calcium-binding protein 1 is a calcium-binding protein discovered in 1999. It has two EF hand motifs and is expressed in neuronal cells in such areas as hippocampus, habenular nucleus of the epithalamus, Purkinje cell layer of the cerebellum, and the amacrine cells and cone bipolar cells of the retina.

Unconventional myosin-Ia is a protein that in humans is encoded by the MYO1A gene.
Forkhead box protein K2 is a protein that in humans is encoded by the FOXK2 gene.

IQ calmodulin-binding motif-containing protein 1 is a protein that in humans is encoded by the IQCB1 gene.
The IQ calmodulin-binding motif is an amino acid sequence motif containing the following sequence:
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.

IQ motif and Sec7 domain 2 is a protein that in humans is encoded by the IQSEC2 gene.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
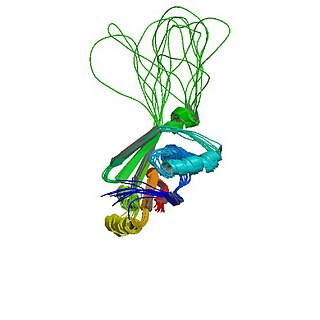
Wilson disease protein (WND), also known as ATP7B protein, is a copper-transporting P-type ATPase which is encoded by the ATP7B gene. The ATP7B protein is located in the trans-Golgi network of the liver and brain and balances the copper level in the body by excreting excess copper into bile and plasma. Genetic disorder of the ATP7B gene may cause Wilson's disease, a disease in which copper accumulates in tissues, leading to neurological or psychiatric issues and liver diseases.
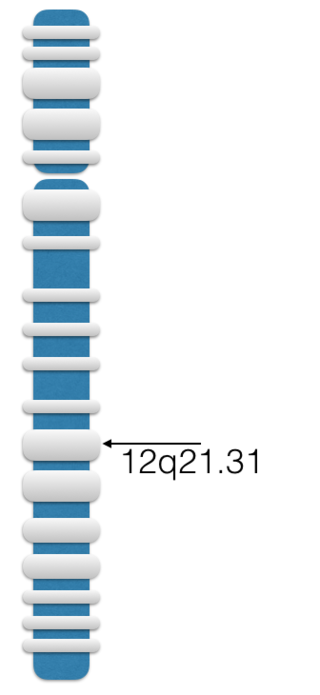
Leucine-rich repeats and IQ motif containing 1 is a protein that in humans is encoded by the LRRIQ1 gene. The protein is likely a nuclear encoding mitochondrial protein and is found in all Metazoans.

LRRIQ3, which is also known as LRRC44, is a protein that in humans is encoded by the LRRIQ3 gene. It is predominantly expressed in the testes, and is linked to a number of diseases.