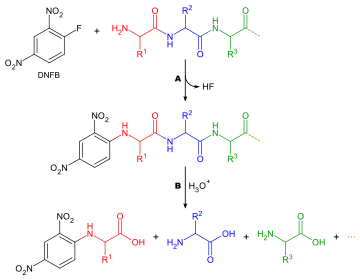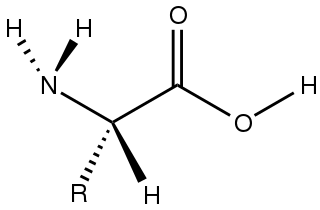
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life.

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.

Cysteine is a semiessential proteinogenic amino acid with the formula HOOC−CH(−NH2)−CH2−SH. The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. Cysteine is chiral, with only L-cysteine being found in nature.

Frederick Sanger was a British biochemist who received the Nobel Prize in Chemistry twice.
In biochemistry, a disulfide refers to a functional group with the structure R−S−S−R′. The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. Persulfide usually refers to R−S−S−H compounds.

Post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes translating mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
Native Chemical Ligation (NCL) is an important extension of the chemical ligation concept for constructing a larger polypeptide chain by the covalent condensation of two or more unprotected peptides segments. Native chemical ligation is the most effective method for synthesizing native or modified proteins of typical size.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a residue, which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo-electron microscopy (cryo-EM) and dual polarisation interferometry, to determine the structure of proteins.
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications. Typically, partial sequencing of a protein provides sufficient information to identify it with reference to databases of protein sequences derived from the conceptual translation of genes.

In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis in living organisms occurs in the opposite direction.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.
Cyanogen bromide is the inorganic compound with the formula (CN)Br or BrCN. It is a colorless solid that is widely used to modify biopolymers, fragment proteins and peptides, and synthesize other compounds. The compound is classified as a pseudohalogen.
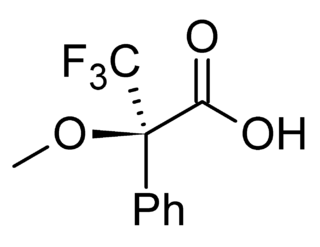
In analytical chemistry, a chiral derivatizing agent (CDA), also known as a chiral resolving reagent, is a derivatization reagent that is a chiral auxiliary used to convert a mixture of enantiomers into diastereomers in order to analyze the quantities of each enantiomer present and determine the optical purity of a sample. Analysis can be conducted by spectroscopy or by chromatography. Some analytical techniques such as HPLC and NMR, in their most commons forms, cannot distinguish enantiomers within a sample, but can distinguish diastereomers. Therefore, converting a mixture of enantiomers to a corresponding mixture of diastereomers can allow analysis. The use of chiral derivatizing agents has declined with the popularization of chiral HPLC. Besides analysis, chiral derivatization is also used for chiral resolution, the actual physical separation of the enantiomers.
Chemical ligation is the chemoselective condensation of unprotected peptide segments enabled by the formation of a non-native bond at the ligation site.
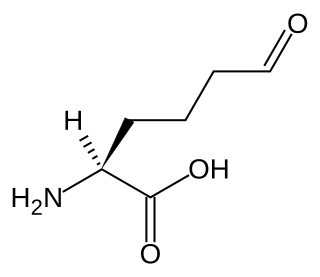
Allysine is a derivative of lysine that features a formyl group in place of the terminal amine. The free amino acid does not exist, but the allysine residue does. It is produced by aerobic oxidation of lysine residues by the enzyme lysyl oxidase. The transformation is an example of a post-translational modification. The semialdehyde form exists in equilibrium with a cyclic derivative.
Bioconjugation is a chemical strategy to form a stable covalent link between two molecules, at least one of which is a biomolecule.
Hydrophobicity scales are values that define the relative hydrophobicity or hydrophilicity of amino acid residues. The more positive the value, the more hydrophobic are the amino acids located in that region of the protein. These scales are commonly used to predict the transmembrane alpha-helices of membrane proteins. When consecutively measuring amino acids of a protein, changes in value indicate attraction of specific protein regions towards the hydrophobic region inside lipid bilayer.
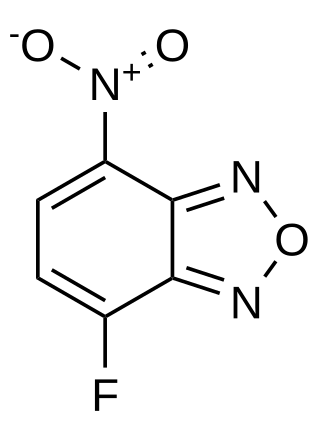
4-Fluoro-7-nitrobenzofurazan (NBD-F) is a fluorogenic, amine labeling dye that is not fluorescent itself, but covalently reacts with secondary or primary amines to form a fluorescently labeled product. It and other fluorogenic benzofurans are used for derivitization in HPLC applications. After the fluorogenic reaction, it can be detected with an excitation wavelength of 470 nm (blue) and an emission wavelength of 530 nm (green), enabling an HPLC limit of detection of 10 fmol.